Abstract
Background
Glioblastoma (GBM) is the most common and malignant gliomas of adults and recur, resulting in death, despite surgery, radiotherapy, and temozolomide-based chemotherapy. There are a few reports on immunotherapy for the mismatch repair (MMR)-deficient GBMs with high tumor mutational burden (TMB). However, the clinicopathological and genetic features of the MMR genes altered in GBMs have not been elucidated yet.
Methods
The authors analyzed targeted next-generation sequencing (NGS) data from 282 (276 primary and 6 recurrent) glioblastomas to evaluate the mutational status of six DNA repair-related genes: MLH1, MSH2, MSH6, PMS2, POLE, and POLD1. Tumors harboring somatic or germline mutations in one or more of these six genes were classified as an MMR gene-altered GBM. The clinicopathologic and molecular characteristics of MMR gene-altered GBMs were compared to those of tumors without MMR gene alterations.
Results
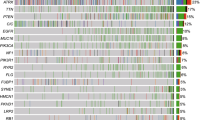
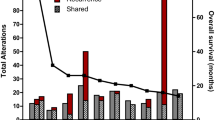
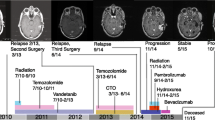
Sixty germline or somatic mutations were identified in 37 cases (35 primary and two recurrent) of GBM. The most frequently mutated genes were MSH6 and POLE. Single nucleotide variants were the most common, followed by frameshift deletions or insertions and approximately 60% of the mutations were germline mutations. Two patients who showed MSH2 (c.2038C > T) and MSH6 (c.1082G > A) mutations had familial colon cancer. The clinical findings were not different between the two groups. However, the presence of MGMT promoter methylation and high tumor mutation burden (TMB) values (> 20) were correlated with MMR gene alterations.
Conclusion
Since MMR-related genes can be found even in primary glioblastoma and are correlated with high TMB and MGMT promoter methylation, MMR genes should be carefully analyzed in NGS study on glioblastomas.



Similar content being viewed by others
Data availability
The data generated and analyzed in this study are provided in the text and the figures, tables, and supplementary tables. Any additional data needed may be requested from the corresponding author.
References
Dolecek TA, Propp JM, Stroup NE, Kruchko C (2012) CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro Oncol 14(Suppl 5):v1-49. https://doi.org/10.1093/neuonc/nos218
Chan TA, Yarchoan M, Jaffee E, Swanton C, Quezada SA, Stenzinger A, Peters S (2019) Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol 30:44–56. https://doi.org/10.1093/annonc/mdy495
McGranahan T, Therkelsen KE, Ahmad S, Nagpal S (2019) Current state of immunotherapy for treatment of glioblastoma. Curr Treat Options Oncol 20:24. https://doi.org/10.1007/s11864-019-0619-4
Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, Ziv E, Culhane AC, Paull EO, Sivakumar IKA, Gentles AJ, Malhotra R, Farshidfar F, Colaprico A, Parker JS, Mose LE, Vo NS, Liu J, Liu Y, Rader J, Dhankani V, Reynolds SM, Bowlby R, Califano A, Cherniack AD, Anastassiou D, Bedognetti D, Mokrab Y, Newman AM, Rao A, Chen K, Krasnitz A, Hu H, Malta TM, Noushmehr H, Pedamallu CS, Bullman S, Ojesina AI, Lamb A, Zhou W, Shen H, Choueiri TK, Weinstein JN, Guinney J, Saltz J, Holt RA, Rabkin CS, Lazar AJ, Serody JS, Demicco EG, Disis ML, Vincent BG, Shmulevich I (2018) The immune landscape of cancer. Immunity 48:812-830.e814. https://doi.org/10.1016/j.immuni.2018.03.023
Indraccolo S, Lombardi G, Fassan M, Pasqualini L, Giunco S, Marcato R, Gasparini A, Candiotto C, Nalio S, Fiduccia P, Fanelli GN, Pambuku A, Della Puppa A, D’Avella D, Bonaldi L, Gardiman MP, Bertorelle R, De Rossi A, Zagonel V (2019) Genetic, epigenetic, and immunologic profiling of mmr-deficient relapsed glioblastoma. Clin Cancer Res 25:1828–1837. https://doi.org/10.1158/1078-0432.ccr-18-1892
Martinez R, Schackert HK, Appelt H, Plaschke J, Baretton G, Schackert G (2005) Low-level microsatellite instability phenotype in sporadic glioblastoma multiforme. J Cancer Res Clin Oncol 131:87–93. https://doi.org/10.1007/s00432-004-0592-5
Mills AM, Liou S, Ford JM, Berek JS, Pai RK, Longacre TA (2014) Lynch syndrome screening should be considered for all patients with newly diagnosed endometrial cancer. Am J Surg Pathol 38:1501–1509. https://doi.org/10.1097/pas.0000000000000321
Geurts-Giele WR, Leenen CH, Dubbink HJ, Meijssen IC, Post E, Sleddens HF, Kuipers EJ, Goverde A, van den Ouweland AM, van Lier MG, Steyerberg EW, van Leerdam ME, Wagner A, Dinjens WN (2014) Somatic aberrations of mismatch repair genes as a cause of microsatellite-unstable cancers. J Pathol 234:548–559. https://doi.org/10.1002/path.4419
Lin KM, Shashidharan M, Thorson AG, Ternent CA, Blatchford GJ, Christensen MA, Watson P, Lemon SJ, Franklin B, Karr B, Lynch J, Lynch HT (1998) Cumulative incidence of colorectal and extracolonic cancers in MLH1 and MSH2 mutation carriers of hereditary nonpolyposis colorectal cancer. J Gastrointest Surg 2:67–71. https://doi.org/10.1016/s1091-255x(98)80105-4
Finocchiaro G, Langella T, Corbetta C, Pellegatta S (2017) Hypermutations in gliomas: a potential immunotherapy target. Discov Med 23:113–120
Chang SC, Lan YT, Lin PC, Yang SH, Lin CH, Liang WY, Chen WS, Jiang JK, Lin JK (2020) Patterns of germline and somatic mutations in 16 genes associated with mismatch repair function or containing tandem repeat sequences. Cancer Med 9:476–486. https://doi.org/10.1002/cam4.2702
Palles C, Cazier JB, Howarth KM, Domingo E, Jones AM, Broderick P, Kemp Z, Spain SL, Guarino E, Salguero I, Sherborne A, Chubb D, Carvajal-Carmona LG, Ma Y, Kaur K, Dobbins S, Barclay E, Gorman M, Martin L, Kovac MB, Humphray S, Lucassen A, Holmes CC, Bentley D, Donnelly P, Taylor J, Petridis C, Roylance R, Sawyer EJ, Kerr DJ, Clark S, Grimes J, Kearsey SE, Thomas HJ, McVean G, Houlston RS, Tomlinson I (2013) Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet 45:136–144. https://doi.org/10.1038/ng.2503
Haraldsdottir S, Hampel H, Tomsic J, Frankel WL, Pearlman R, de la Chapelle A, Pritchard CC (2014) Colon and endometrial cancers with mismatch repair deficiency can arise from somatic, rather than germline, mutations. Gastroenterology 147:1308-1316.e1301. https://doi.org/10.1053/j.gastro.2014.08.041
Hodges TR, Ott M, Xiu J, Gatalica Z, Swensen J, Zhou S, Huse JT, de Groot J, Li S, Overwijk WW, Spetzler D, Heimberger AB (2017) Mutational burden, immune checkpoint expression, and mismatch repair in glioma: implications for immune checkpoint immunotherapy. Neuro Oncol 19:1047–1057. https://doi.org/10.1093/neuonc/nox026
Liu Y, Chen L, Zhang S, Shu Y, Qi Q, Zhu M, Peng Y, Ling Y (2020) Somatic mutations in genes associated with mismatch repair predict survival in patients with metastatic cancer receiving immune checkpoint inhibitors. Oncol Lett 20:27. https://doi.org/10.3892/ol.2020.11888
Comprehensive genomic characterization defines human glioblastoma genes and core pathways (2008). Nature 455:1061–1068. https://doi.org/https://doi.org/10.1038/nature07385
Cahill DP, Levine KK, Betensky RA, Codd PJ, Romany CA, Reavie LB, Batchelor TT, Futreal PA, Stratton MR, Curry WT, Iafrate AJ, Louis DN (2007) Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res 13:2038–2045. https://doi.org/10.1158/1078-0432.Ccr-06-2149
Yao J, Gong Y, Zhao W, Han Z, Guo S, Liu H, Peng X, Xiao W, Li Y, Dang S, Liu G, Li L, Huang T, Chen S, Song L (2019) Comprehensive analysis of POLE and POLD1 Gene Variations identifies cancer patients potentially benefit from immunotherapy in Chinese population. Scientific Reports 9:15767. https://doi.org/10.1038/s41598-019-52414-z
Kim JE, Chun SM, Hong YS, Kim KP, Kim SY, Kim J, Sung CO, Cho EJ, Kim TW, Jang SJ (2019) Mutation Burden and I Index for Detection of Microsatellite Instability in Colorectal Cancer by Targeted Next-Generation Sequencing. J Mol Diagn 21:241–250. https://doi.org/10.1016/j.jmoldx.2018.09.005
Lommatzsch M, Bratke K, Stoll P (2018) Neoadjuvant PD-1 Blockade in Resectable Lung Cancer. N Engl J Med 379:e14. https://doi.org/10.1056/NEJMc1808251
Necchi A, Anichini A, Raggi D, Briganti A, Massa S, Luciano R, Colecchia M, Giannatempo P, Mortarini R, Bianchi M, Fare E, Monopoli F, Colombo R, Gallina A, Salonia A, Messina A, Ali SM, Madison R, Ross JS, Chung JH, Salvioni R, Mariani L, Montorsi F (2018) Pembrolizumab as Neoadjuvant Therapy Before Radical Cystectomy in Patients With Muscle-Invasive Urothelial Bladder Carcinoma (PURE-01) An Open-Label Single-Arm. J Clin Oncol, Phase II Study. https://doi.org/10.1200/jco.18.01148
McCord M, Steffens A, Javier R, Kam KL, McCortney K, Horbinski C (2020) The efficacy of DNA mismatch repair enzyme immunohistochemistry as a screening test for hypermutated gliomas. Acta Neuropathol Commun 8:15. https://doi.org/10.1186/s40478-020-0892-2
Yip S, Miao J, Cahill DP, Iafrate AJ, Aldape K, Nutt CL, Louis DN (2009) MSH6 mutations arise in glioblastomas during temozolomide therapy and mediate temozolomide resistance. Clin Cancer Res 15:4622–4629. https://doi.org/10.1158/1078-0432.Ccr-08-3012
Xie C, Sheng H, Zhang N, Li S, Wei X, Zheng X (2016) Association of MSH6 mutation with glioma susceptibility, drug resistance and progression. Mol Clin Oncol 5:236–240. https://doi.org/10.3892/mco.2016.907
Hunter C, Smith R, Cahill DP, Stephens P, Stevens C, Teague J, Greenman C, Edkins S, Bignell G, Davies H, O’Meara S, Parker A, Avis T, Barthorpe S, Brackenbury L, Buck G, Butler A, Clements J, Cole J, Dicks E, Forbes S, Gorton M, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, Kosmidou V, Laman R, Lugg R, Menzies A, Perry J, Petty R, Raine K, Richardson D, Shepherd R, Small A, Solomon H, Tofts C, Varian J, West S, Widaa S, Yates A, Easton DF, Riggins G, Roy JE, Levine KK, Mueller W, Batchelor TT, Louis DN, Stratton MR, Futreal PA, Wooster R (2006) A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res 66:3987–3991. https://doi.org/10.1158/0008-5472.Can-06-0127
Luchini C, Bibeau F, Ligtenberg MJL, Singh N, Nottegar A, Bosse T, Miller R, Riaz N, Douillard JY, Andre F, Scarpa A (2019) ESMO recommendations on microsatellite instability testing for immunotherapy in cancer, and its relationship with PD-1/PD-L1 expression and tumour mutational burden: a systematic review-based approach. Ann Oncol 30:1232–1243. https://doi.org/10.1093/annonc/mdz116
Southey MC, Jenkins MA, Mead L, Whitty J, Trivett M, Tesoriero AA, Smith LD, Jennings K, Grubb G, Royce SG, Walsh MD, Barker MA, Young JP, Jass JR, St John DJB, Macrae FA, Giles GG, Hopper JL (2005) Use of Molecular Tumor Characteristics to Prioritize Mismatch Repair Gene Testing in Early-Onset Colorectal Cancer. J Clin Oncol 23:6524–6532. https://doi.org/10.1200/JCO.2005.04.671
Engel KB, Moore HM (2011) Effects of preanalytical variables on the detection of proteins by immunohistochemistry in formalin-fixed, paraffin-embedded tissue. Arch Pathol Lab Med 135:537–543. https://doi.org/10.1043/2010-0702-rair.1
Shia J (2008) Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome. Part I. The utility of immunohistochemistry. J Mol Diagn 10:293–300. https://doi.org/10.2353/jmoldx.2008.080031
Baudrin LG, Deleuze JF, How-Kit A (2018) Molecular and Computational Methods for the Detection of Microsatellite Instability in Cancer. Front Oncol 8:621. https://doi.org/10.3389/fonc.2018.00621
Waalkes A, Smith N, Penewit K, Hempelmann J, Konnick EQ, Hause RJ, Pritchard CC, Salipante SJ (2018) Accurate Pan-Cancer Molecular Diagnosis of Microsatellite Instability by Single-Molecule Molecular Inversion Probe Capture and High-Throughput Sequencing. Clin Chem 64:950–958. https://doi.org/10.1373/clinchem.2017.285981
Bartley AN, Luthra R, Saraiya DS, Urbauer DL, Broaddus RR (2012) Identification of Cancer Patients with Lynch Syndrome: Clinically Significant Discordances and Problems in Tissue-Based Mismatch Repair Testing. Cancer Prevention Research 5:320. https://doi.org/10.1158/1940-6207.CAPR-11-0288
Forsström LM, Sumi K, Mäkinen MJ, Oh JE, Herva R, Kleihues P, Ohgaki H, Aaltonen LA (2017) Germline MSH6 Mutation in a Patient With Two Independent Primary Glioblastomas. J Neuropathol Exp Neurol 76:848–853. https://doi.org/10.1093/jnen/nlx066
Maxwell JA, Johnson SP, McLendon RE, Lister DW, Horne KS, Rasheed A, Quinn JA, Ali-Osman F, Friedman AH, Modrich PL, Bigner DD, Friedman HS (2008) Mismatch repair deficiency does not mediate clinical resistance to temozolomide in malignant glioma. Clinical cancer research : an official journal of the American Association for Cancer Research 14:4859–4868. https://doi.org/10.1158/1078-0432.CCR-07-4807
Salipante SJ, Scroggins SM, Hampel HL, Turner EH, Pritchard CC (2014) Microsatellite instability detection by next generation sequencing. Clin Chem 60:1192–1199. https://doi.org/10.1373/clinchem.2014.223677
Niu B, Ye K, Zhang Q, Lu C, Xie M, McLellan MD, Wendl MC, Ding L (2013) MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics 30:1015–1016. https://doi.org/10.1093/bioinformatics/btt755
Kautto EA, Bonneville R, Miya J, Yu L, Krook MA, Reeser JW, Roychowdhury S (2016) Performance evaluation for rapid detection of pan-cancer microsatellite instability with MANTIS. Oncotarget 8(5):7452–7463
Haraldsdottir S (2017) Microsatellite Instability Testing Using Next-Generation Sequencing Data and Therapy Implications. JCO Precision Oncology 1:1–4. https://doi.org/10.1200/PO.17.00189
Louis DN, Ohgaki, H., Wiestler, O.D., Cavenee, W.K. (2016) WHO Classification of Tumours of the Central Nervous System, Revised. Fourth Edition.
Park S, Lee H, Lee B, Lee SH, Sun JM, Park WY, Ahn JS, Ahn MJ, Park K (2019) DNA Damage Response and Repair Pathway Alteration and Its Association With Tumor Mutation Burden and Platinum-Based Chemotherapy in SCLC. J Thorac Oncol 14:1640–1650. https://doi.org/10.1016/j.jtho.2019.05.014
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. https://doi.org/10.1126/scisignal.2004088
Lee S, Seo J, Park J, Nam JY, Choi A, Ignatius JS, Bjornson RD, Chae JH, Jang IJ, Lee S, Park WY, Baek D, Choi M (2017) Korean Variant Archive (KOVA): a reference database of genetic variations in the Korean population. Sci Rep 7:4287. https://doi.org/10.1038/s41598-017-04642-4
Johnson A, Severson E, Gay L, Vergilio JA, Elvin J, Suh J, Daniel S, Covert M, Frampton GM, Hsu S, Lesser GJ, Stogner-Underwood K, Mott RT, Rush SZ, Stanke JJ, Dahiya S, Sun J, Reddy P, Chalmers ZR, Erlich R, Chudnovsky Y, Fabrizio D, Schrock AB, Ali S, Miller V, Stephens PJ, Ross J, Crawford JR, Ramkissoon SH (2017) Comprehensive Genomic Profiling of 282 Pediatric Low- and High-Grade Gliomas Reveals Genomic Drivers, Tumor Mutational Burden, and Hypermutation Signatures. Oncologist 22:1478–1490. https://doi.org/10.1634/theoncologist.2017-0242
Bonneville R, Krook MA, Kautto EA, Miya J, Wing MR, Chen H-Z, Reeser JW, Yu L, Roychowdhury S (2017) Landscape of Microsatellite Instability Across 39 Cancer Types. JCO Precision Oncology. https://doi.org/10.1200/po.17.00073
Daniel P, Sabri S, Chaddad A, Meehan B, Jean-Claude B, Rak J, Abdulkarim BS (2019) Temozolomide Induced Hypermutation in Glioma: Evolutionary Mechanisms and Therapeutic Opportunities. Front Oncol 9:41. https://doi.org/10.3389/fonc.2019.00041
Sa JK, Choi SW, Zhao J, Lee Y, Zhang J, Kong DS, Choi JW, Seol HJ, Lee JI, Iavarone A, Rabadan R, Nam DH (2019) Hypermutagenesis in untreated adult gliomas due to inherited mismatch mutations. Int J Cancer 144:3023–3030. https://doi.org/10.1002/ijc.32054
Stelloo E, Jansen AML, Osse EM, Nout RA, Creutzberg CL, Ruano D, Church DN, Morreau H, Smit V, van Wezel T, Bosse T (2017) Practical guidance for mismatch repair-deficiency testing in endometrial cancer. Ann Oncol 28:96–102. https://doi.org/10.1093/annonc/mdw542
Yuan L, Chi Y, Chen W, Chen X, Wei P, Sheng W, Zhou X, Shi D (2015) Immunohistochemistry and microsatellite instability analysis in molecular subtyping of colorectal carcinoma based on mismatch repair competency. Int J Clin Exp Med 8:20988–21000
Lee JH, Cragun D, Thompson Z, Coppola D, Nicosia SV, Akbari M, Zhang S, McLaughlin J, Narod S, Schildkraut J, Sellers TA, Pal T (2014) Association between IHC and MSI testing to identify mismatch repair-deficient patients with ovarian cancer. Genet Test Mol Biomarkers 18:229–235. https://doi.org/10.1089/gtmb.2013.0393
Chalmers ZR, Connelly CF, Fabrizio D, Gay L, Ali SM, Ennis R, Schrock A, Campbell B, Shlien A, Chmielecki J, Huang F, He Y, Sun J, Tabori U, Kennedy M, Lieber DS, Roels S, White J, Otto GA, Ross JS, Garraway L, Miller VA, Stephens PJ, Frampton GM (2017) Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 9:34. https://doi.org/10.1186/s13073-017-0424-2
Kroeze LI, de Voer RM, Kamping EJ, von Rhein D, Jansen EAM, Hermsen MJW, Barberis MCP, Botling J, Garrido-Martin EM, Haller F, Lacroix L, Maes B, Merkelbach-Bruse S, Pestinger V, Pfarr N, Stenzinger A, van den Heuvel MM, Grunberg K, Ligtenberg MJL (2020) Evaluation of a Hybrid Capture-Based Pan-Cancer Panel for Analysis of Treatment Stratifying Oncogenic Aberrations and Processes. J Mol Diagn. https://doi.org/10.1016/j.jmoldx.2020.02.009
Vanderwalde A, Spetzler D, Xiao N, Gatalica Z, Marshall J (2018) Microsatellite instability status determined by next-generation sequencing and compared with PD-L1 and tumor mutational burden in 11,348 patients. Cancer Med 7:746–756. https://doi.org/10.1002/cam4.1372
Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V, Stephens PJ, Daniels GA, Kurzrock R (2017) Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther 16:2598–2608. https://doi.org/10.1158/1535-7163.mct-17-0386
Medikonda R, Dunn G, Rahman M, Fecci P, Lim M (2020) A review of glioblastoma immunotherapy. J Neurooncol. https://doi.org/10.1007/s11060-020-03448-1
Hunter C, Smith R, Cahill DP, Stephens P, Stevens C, Teague J, Greenman C, Edkins S, Bignell G, Davies H, O’Meara S, Parker A, Avis T, Barthorpe S, Brackenbury L, Buck G, Butler A, Clements J, Cole J, Dicks E, Forbes S, Gorton M, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, Kosmidou V, Laman R, Lugg R, Menzies A, Perry J, Petty R, Raine K, Richardson D, Shepherd R, Small A, Solomon H, Tofts C, Varian J, West S, Widaa S, Yates A, Easton DF, Riggins G, Roy JE, Levine KK, Mueller W, Batchelor TT, Louis DN, Stratton MR, Futreal PA, Wooster R (2006) A Hypermutation Phenotype and Somatic <em>MSH6</em> mutations in recurrent human malignant gliomas after alkylator chemotherapy. Can Res 66:3987–3991. https://doi.org/10.1158/0008-5472.Can-06-0127
Choi S, Yu Y, Grimmer MR, Wahl M, Chang SM, Costello JF (2018) Temozolomide-associated hypermutation in gliomas. Neuro-Oncology 20:1300–1309. https://doi.org/10.1093/neuonc/noy016
Bouffet E, Larouche V, Campbell BB, Merico D, de Borja R, Aronson M, Durno C, Krueger J, Cabric V, Ramaswamy V, Zhukova N, Mason G, Farah R, Afzal S, Yalon M, Rechavi G, Magimairajan V, Walsh MF, Constantini S, Dvir R, Elhasid R, Reddy A, Osborn M, Sullivan M, Hansford J, Dodgshun A, Klauber-Demore N, Peterson L, Patel S, Lindhorst S, Atkinson J, Cohen Z, Laframboise R, Dirks P, Taylor M, Malkin D, Albrecht S, Dudley RW, Jabado N, Hawkins CE, Shlien A, Tabori U (2016) Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J Clin Oncol 34:2206–2211. https://doi.org/10.1200/jco.2016.66.6552
Johanns TM, Miller CA, Dorward IG, Tsien C, Chang E, Perry A, Uppaluri R, Ferguson C, Schmidt RE, Dahiya S, Ansstas G, Mardis ER, Dunn GP (2016) Immunogenomics of hypermutated glioblastoma: a patient with germline POLE deficiency treated with checkpoint blockade immunotherapy. Cancer Discov 6:1230–1236. https://doi.org/10.1158/2159-8290.Cd-16-0575
Ahmad H, Fadul CE, Schiff D, Purow B (2019) Checkpoint inhibitor failure in hypermutated and mismatch repair-mutated recurrent high-grade gliomas. Neuro-oncology practice 6:424–427. https://doi.org/10.1093/nop/npz016
Funding
None.
Author information
Authors and Affiliations
Contributions
Contribution to design: YLS, YAC. Contribution to analysis and interpretation of the data: YAC, DGK, BL, JHS. YAC and YLS wrote the manuscript. All authors were involved in writing this manuscript and read and approved the final version.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare no conflicts of interest.
Ethical approval
The Institutional Review Board of Samsung Medical Center approved this study and waived the requirement for informed consent (IRB Number: 2020–04-011).
Consent for publication
No individual patient data were reported in this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cho, Y.A., Kim, D., Lee, B. et al. Incidence, clinicopathologic, and genetic characteristics of mismatch repair gene-mutated glioblastomas. J Neurooncol 153, 43–53 (2021). https://doi.org/10.1007/s11060-021-03710-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-021-03710-0