Abstract
Purpose
Ecotropic viral integration site-1 (EVI1) is a transcription factor that contributes to the unfavorable prognosis of leukemia, some epithelial cancers, and glial tumors. However, the biological function of EVI1 in glioblastoma multiforme (GBM) remains unclear. Based on microarray experiments, EVI1 has been reported to regulate epidermal growth factor receptor (EGFR) transcription. Signal transduction via EGFR plays an essential role in glioblastoma. Therefore, we performed this study to clarify the importance of EVI1 in GBM by focusing on the regulatory mechanism between EVI1 and EGFR transcription.
Methods
We performed immunohistochemical staining and analyzed the EVI1-expression in glioma tissue. To determine the relationship between EVI1 and EGFR, we induced siRNA-mediated knockdown of EVI1 in GBM cell lines. To investigate the region that was essential for the EVI1 regulation of EGFR expression, we conducted promoter reporter assays. We performed WST-8 assay to investigate whether EVI1 affected on the proliferation of GBM cells or not.
Results
It was observed that 22% of GBM tissues had over 33% of tumor cells expressing EVI1, whereas no lower-grade glioma tissue had over 33% by immunohistochemistry. In A172 and YKG1 cells, the expression levels of EGFR and EVI1 correlated. Analysis of the EGFR promoter region revealed that the EGFR promoter (from − 377 to − 266 bp) was essential for the EVI regulation of EGFR expression. We showed that EVI1 influenced the proliferation of A172 and YKG1 cells.
Conclusion
This is the first study reporting the regulation of EGFR transcription by EVI1 in GBM cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In 1988, ecotropic viral integration site-1 (EVI1) was originally identified as a common site of retroviral integration in murine myeloid tumors [1, 2]. The human EVI1 is located on chromosome 3q26 and produces a transcription factor that contains two DNA-binding zinc finger domains: one binds to a GATA-like consensus motif and the other binds to a v-ets erythroblastosis virus E26 oncogene homolog (ETS)-like motif [3, 4]. While regulating target gene expression, EVI1 interacts with transcription coregulators such as the C-terminal binding domain (CtBP), cAMP-responsive element-binding protein-binding protein (CBP), and p300/CBP-associated factor (P/CAF) [5]. EVI1 represses transforming growth factor-beta (TGF-β) signaling and activates PI3K/Akt/mTOR signaling [6, 7]. Elevated EVI1 expression is an unfavorable prognostic factor in human acute myeloid leukemia and some solid cancers [8,9,10,11,12].
EVI1 is closely related to embryonic neural development. In one study, the EVI1 homozygous mutant mouse embryos that died at 10.5 day post-coitus had a defect in the cranial ganglia and developed failure of the peripheral nervous system [13]. EVI1 is related to the Notch signaling pathway, which is important for cell-fate specification in a developing mammalian nervous system [5, 6]. This evidence showed that EVI1 is a key molecule in neurogenesis. Further, EVI1 has been reported to contribute to worsening of glial tumors [7, 8], suggesting that it might have an oncogenic role in glioma genesis. However, studies aiming to investigate EVI1 function in glioma have been few.
Recently, through cDNA microarray Gene Chips experiments, Chapeau et al. revealed that EVI1 regulated 621 cancer-associated genes in Hela and SKOV3 cells [9]. Among these EVI1 target genes, we were interested in epidermal growth factor receptor (EGFR). Signal transduction via the tyrosine kinase receptor EGFR contributes to cell proliferation, apoptosis inhibition, and angiogenesis in cancer cells. EGFR is the most important downstream target gene of the Notch signaling pathway [10, 11]. Since amplification and/or mutations in EGFR represent a genetic abnormality in primary glioblastoma multiforme (GBM), EGFR has been regarded as a pivotal target for GBM studies.
Therefore, we presumed that EVI1 might have a significant role in GBM by regulating EGFR gene expression.
Materials and methods
Microarray data
We downloaded the four GEO series (GSE2223, GSE4271, GSE23806, and GSE43378) from the GEO database in NCBI (https://www.ncbi.nlm.nih.gov/geo/). Here we chose 29 GBM samples, except gliosarcoma, from GSE2223. In the same way, we chose 76 GBM samples from GSE4271, chose 32 conventional GBM cell line samples from GSE23806 and chose 32 GBM samples from GSE43378. We used pairs of probe-sets of MECOM and EGFR to evaluate the corresponding expression levels, demonstrated by scatter plot. (Supplementary Fig. 1).
Sample collection
This study was approved by the Ethics Committee of the University of Miyazaki Hospital, Miyazaki, Japan. All patients and their families gave informed consent for the use of the resected tissues. In this study, we included patients who had infiltrative glioma with histological grades II and III (lower-grade glioma, n = 27; 15 men and 12 women; median age, 41 years) or grade IV (GBM, n = 37; 27 men and 10 women; median age, 67 years). We used eight normal brain tissues as normal controls. Two or more pathologists examined all tumor tissue sections according to the WHO criteria. The tissue samples were obtained from tumors resected at the Department of Neurosurgery, University of Miyazaki Hospital between May 2014 and January 2017.
Immunohistochemical staining and evaluation of the paraffin-embedded tissues
To evaluate the expression of EVI1 using immunohistochemistry, we used the diluted EVI1 antibody (1:500, Cell Signaling Technology, #2593). To evaluate the expression of EGFR using immunohistochemistry, we used the diluted EGFR antibody (1:200, Leica microsystems, NCL-EGFR). The primary antibody was detected using the Dako EnVision™ + system—HRP Labeled Polymer (anti-rabbit) (Dako, K4002) and the Dako EnVision™ + system–HRP Labeled Polymer (anti-mouse) (Dako, K4000). The slides were counterstained using Meyer’s hematoxylin. Without knowledge on the clinical information, we photographed three regions of high cellularity in each tissue slide using a BZ-9000 microscope (Keyence, Osaka, Japan). The numbers of both EVI1-positive cells and cells in total were calculated using BZ-II Analyzer version 1.42 and BZ-II Dynamic Cell Count version 1.01 (Keyence). The mean values were defined as the EVI1-positive cell rate. The scores of the EVI1-positive tumor cells were defined as (−) if less than 5% (+) if 5–33% (++) if 33–50%, and (+++) if more than 50%. The threshold values were defined based on those described in the previous study by Hou et al. [8]
PCR and real-time quantitative PCR
Total RNA was isolated using the RNeasy Mini Kit, according to the manufacturer’s instructions (Qiagen, No.74104). Total RNA in each sample was reverse transcribed using the Super Script VILO cDNA synthesis kit (Invitrogen, 11754-050). Quantitative PCR was performed on the StepOnePlus (Applied bio systems) using THUNDERBIRD SYBR qPCR Mix (TOYOBO, QPS- 201). The primers used in this study are as described in Supplementary Fig. 2.
EVI1 RNA interference
The final RNA concentration was 16.6 nM. The siRNA sequences targeting human EVI1 were as described in Supplementary Fig. 2 [12]. The universal negative control siRNA was purchased from NIPPON GENE (NIPPON GENE CO. Ltd. Toyama, Japan).
Western blotting
We used the primary antibody (EGFR, Cell Signaling Technology_#4267_1:1000 and b-Actin, SIGMA-ALDRICH_A5441_1:5000). The secondary antibody was applied and the signal was visualized on ImageQuant LAS 4000 (GE Healthcare) using the Lumi-Light plus Western Blotting Substrate (Roche, 12015196001).
Constructs
All constructs that we used in this study are described in details in Electrical Supplementary Material 1.
Luciferase assay
We performed the dual luciferase assays according to the manufacturer’s instructions (Promega, E1910).
Cell proliferation assay
Cell proliferation assays were performed with a Cell Counting Kit-8 (Dojindo, Japan) according to the manufacturer’s protocol.
Statistical analysis
The correlations between the expression levels of EVI1 and EGFR (GSE2223, GSE4271, GSE23806, and GSE43378) were calculated using Spearman’s rank correlation coefficient (Supplementary Fig. 1). The difference in the survival curves was analyzed by the log-rank test (Fig. 1c). We compared the EVI1-mRNA levels of the target genes between the GBM cell lines and the normal brain tissues using Mann–Whitney U-test (Fig. 2a). All statistical analyses were performed using the Student’s t test (Figs. 2, 3, 4). We used EasyR software for statistical analysis. P values < 0.05 were considered significant.
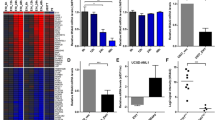
Immunohistochemical staining for EVI1 in glioma tissue. a Representative HE staining (× 400) (i, iii) and EVI1 immunohistochemistry images (ii, iv) of GBM tissues; (i) and (ii) are GBM tissues expressing high levels of EVI1, whereas (iii) and (iv) are GBM tissues expressing low levels of EVI1. The scale bars represent 100 μm. b Representative immunohistochemistry images (× 200) of GBM tissues; (i) and (ii) are GBM tissues expressing high levels of EVI1, whereas (iii) and (iv) are GBM tissues expressing low levels of EVI1. (i, iii) were EVI1 immunohistochemistry, whereas (ii, iv) were EGFR immunohistochemistry. The scale bars represent 100 μm. c Log-rank test for the survival curves stratified by EVI1 expression in 37 GBM tissues shows a worse overall survival rate in patients with high EVI1 expression (EVI1-positive cell rate of ≥ 33%; n = 8) than in those with low EVI1 expression (EVI1-positive cell rate of < 33%; n = 29) (P = 0.0109)
a The expression levels of EVI1 in GBM cell lines. The mRNA expressions of EVI1 (left panel) and protein of EVI1 (right panel) in 7 GBM cell lines (A-172, LN-18, LN-229, T98G, U-87MG, U-251MG, and YKG1) are shown. In the western blot analysis of EVI1 in GBM cell lines and normal brain tissues, the beta-actin was used as the loading control. Triplicates were analyzed and results were presented as changes in fold, relative to the A172-EVI1 expression level. The error bars represent the standard deviation. (P < 0.05) (The representative data was shown in Supplementary Fig. 4) The graphs show the integrated density measured by Image J and normalized to beta-actin. b siRNA-mediated EVI1 knockdown and the expression level of EGFR in GBM cells. (A172, YKG1) The mRNA expressions of EVI1 and EGFR were normalized to 18S mRNA (left panel, A172 and right panel, YKG1). The black bars represent negative control, the white bars represent EVI1 knock down (siEVI1_B) and the gray bars represent EVI1 knock down (siEVI1_C). All assays were performed in triplicate. The results were presented as changes in fold, relative to the negative control. Quantitative PCR data were presented as mean ± SD mRNA expression, *P < 0.05. c The relative mRNA expressions of EVI1 (left panel) and EGFR (right panel) in the A172 cells that underwent siRNA transfection. EGFR-mRNA levels decreased from 24 to 96 h. The highest difference in EGFR-mRNA expression level relative to the negative control was observed at 96 h after transfection. d Western blot analysis of EGFR in GBM cells (left panel, A172 and right panel, YKG-1). The beta-actin was the loading control. Triplicates were analyzed and results were presented as changes in fold, relative to the negative control. The graphs show the integrated density measured by Image J and normalized to beta-actin. The error bars represent the standard deviation. (P < 0.02)
The importance of ERR for EVI1 regulation of EGFR transcription. All dual luciferase assays in the GBM cells were performed in triplicate. The values and error bars depict the mean values ± SD. a Various length fragments of the EGFR promoter region were inserted upstream of the luciferase gene in the reporter plasmid pGL4.10 [luc2]. The relative luciferase activity was normalized after transfection in U-87MG. A pGL4.10 [luc2] plasmid was used as control (pGL4-empty vector). **P < 0.02. b The relative luciferase activities after transfection of the pGL4-377 plasmid with the EVI1 overexpression vector (pCMV26-EVI1) or control vector (pCMV26-control). The overexpression of EVI1 protein in GBM cells (U87MG, YKG1) was validated by western blot analysis (Supplementary Fig. 7). This experiment was performed using the GBM cell lines LN229, YKG-1, U-251MG, U-87MG, and A-172, **P < 0.02. c The relative luciferase activities in U-87MG after transfection of the luciferase reporter plasmids, with various lengths of EGFR promoter fragments integrated. Each plasmid was transfected with the EVI1 overexpression vector (pMXs-EVI1; black bars) or the control vector (pMXs-control; gray bars). pGL265 with the control vector was used as control. All luciferase reporter assays were performed in quadruplicate. The values and error bars depict the mean ± SD, *P < 0.05. d The panel shows the sequences of ERR (EVI1 regulatory region). The sequences are numbered according to their positions, relative to the site of translation start (+ 1 at ATG). The two boxed sequences represent ERR1 (− 377 bp to − 336 bp) and ERR2 (− 335 bp to − 266 bp) sites. The mutation regions that we used in the EGFR promoter mutant luciferase assay are shown in this schema. e The EGFR promoter regions (− 377 bp to − 63 bp upstream of the translational starting site), which contain mutations (X) in the putative EVI1 binding sites, are inserted upstream of the luciferase gene in the reporter plasmid pGL4.10 [luc2]. The numbers placed on the X mark represent a region from the site of the translational start of the EGFR promoter. Each plasmid is transfected with the EVI1 overexpression vector (pMXs-EVI1, black bars) or the control vector (pMXs-control, gray bars) into U-87MG. **P < 0.02. f Enhancement of the promoter activity of EGFR by EVI1 via the C-terminal DNA-binding domain. The structures of the wild-type EVI1 (EVI1 wt) and the mutant EVI1 containing a deletion of the C-terminal DNA-binding domains (EVI1 Δ8-10) are depicted. U-87MG, U-251MG, and YKG-1 cells were co-transfected with the EGFR reporter vector (pGL4-377), pRL-TK, and each EVI1 overexpression vector (pCMV26-EVI1/ pCMV26-EVI1Δ8-10). pCMV26-control was used as control. All luciferase reporter assays were performed in triplicate. The values and error bars depict the mean ± SD, *P < 0.05
The cell proliferation curves of the A172 and YKG-1 cells transduced with siRNA against EVI1 (siEVI1_B and siEVI1_C), compared with those of the negative control (siRNA-control). On day 0, the cells were transfected with siRNA. Cell counting was carried out at 24, 48, and 72 h after siRNA transfection. Data shown are the mean ± SD of the quadruplicate analyses. Statistical significance was determined by the Student’s t test, *P < 0.05
Results
Positive correlation between the expression levels of EVI1 and EGFR in GBM
To investigate whether EVI1 was associated with EGFR expression in GBM, we referred to GEO data and calculated the correlation coefficients of the expression levels of EVI1 and EGFR. A positive correlation was found between EVI1 and EGFR expressions in each of the GBM series, including GSE2223 (r = 0.429–0.51); GSE4271 (r = 0.379–0.462); GSE23806 (r = 0.445–0.606); and GSE43378 (r = 0.461–0.551) (Supplementary Fig. 1).
The EVI1 expression level was relatively high in GBM tissue and might contribute to worse prognosis
To clarify the role of EVI1 in glioma genesis, we performed immunohistochemical staining and analyzed the EVI1-positive cell rates in 37 GBM and 27 lower-grade glioma (LGG) tissue samples. EVI1 was detected in the nuclei of the glioma cells, and EVI1-positive tumor cells were observed diffusely in glioma tissue. We observed the heterogeneity of nuclei at the point of staining intensity (Fig. 1a). In addition, the cases those were stained well by EVI1 were also stained well by EGFR (Fig. 1b).
On examination of the 37 GBM tissue samples, the positive rate was 0–5% in 15 cases, 5–33% in 14 cases, 33–50% in six cases, and over 50% in two cases. On the other hand, among the 27 LGG tissue samples, the positive rate was 0–5% in 13 cases and 5–33% in 14 cases; none of the cases had over 33% positive rate (Table 1). Because no LGG tissues had over 33% of tumor cells expressing EVI1, we chose the 33% as the cut-off level in the survival curve. Of the 37 GBM cases, eight cases that had over 33% EVI1-positive rate showed significantly worse prognosis (P = 0.0109). The one-year survival rate was 12.5% for the cases with over 33% positive rate and 62.2% for the cases with 0–33% positive rate (Fig. 1c). On the other hand, we examined the relation between the level of EGFR-mRNA expression and post-operative days in these patients with GBM. However, the result showed no significant difference (Supplementary Fig. 3).
Among the 37 GBM tissue samples, the positive rate was over 33% in eight cases. However, among the 27 LGG tissue samples, none showed a positive rate of over 33%. The scores of the EVI1-positive tumor cells were defined as (−) if less than 5% (+) if 5% to 33% (++) if 33% to 50%, and (+++) if more than 50%.
The EVI1 expression levels were more varied in the GBM cell lines than in the normal brain tissue samples
To investigate the expression levels of EVI1 in various GBM cell lines, we performed quantitative PCR and western blotting using seven GBM cell lines (i.e., A-172, LN-18, LN-229, T98G, U-87MG, U-251MG, and YKG-1) and compared them with those in normal brain tissues (i.e., NB-1, NB-2, NB-3, NB-4, NB-5, NB-6, NB-7, and NB-8). The levels of EVI1-mRNA in the GBM cell lines varied and showed no significant difference; (P = 0.052) (Fig. 2a, left panel), however, the protein levels of EVI1 in GBM cell lines were higher compared with those in normal brain tissues (P < 0.05) (Fig. 2a, right panel, The representative data was shown in Supplementary Fig. 4).
siRNA-mediated EVI1 knockdown decreased the expression levels of EGFR
To determine the regulatory mechanism between EVI1 and EGFR, we induced knockdown of EVI1 in GBM cells because EVI1 is a transcription factor. First, we induced siRNA-mediated knock down in A172, LN18, LN229, T98G, U87MG, U251MG, and YKG1. As a result, the EVI1-mRNA levels in these GBM cell lines were reduced. In addition, we observed reduction in EGFR-mRNA and EGFR protein expression in response to the decrease in EVI1 mRNA (Supplementary Figs. 5, 6). Among these GBM cell lines, the differences in EGFR-mRNA between knock down group and control group were bigger in A172 and YKG1 than other GBM cell lines. Thus, we chose these two cell lines for siRNA-mediated EVI1 knock down experiments.
Transfection with the siRNAs achieved > 75% decrease in EVI1 mRNA expression in the A172 and YKG-1 cells. In the A172 cells, EGFR-mRNA expression decreased by 35% and 45% after transfection with siEVI1_B and siEVI1_C, respectively, whereas in the YKG1 cells, the expression decreased by 49% and 45% after transfection with siEVI1_B and siEVI1_C, respectively (Fig. 2b).
In the A172 cells transfected with siEVI1_C, reduced EVI1 and EGFR-mRNA levels were observed at 24, 48, 72, and 96 h; the largest difference in the EGFR-mRNA expression level, relative to the negative control, was observed at 96 h after transfection (66% reduction) (Fig. 2c). Therefore, we conducted western blot assay 96 h after transfection and found that the EGFR protein expression in GBM cells decreased after treatment with the siRNAs against EVI1, compared with the effects in the negative control. In the A172 cells, EGFR protein expression decreased by 65% and 79% after transfection with siEVI1_B and siEVI1_C, respectively, whereas in the YKG1 cells, the expression decreased by 27% and 47% after transfection with siEVI1_B and siEVI1_C, respectively (Fig. 2d).
The importance of the EVI1 regulatory region in the EVI1 regulation of EGFR transcription
To clarify the molecular mechanisms underlying the transcriptional regulation of EGFR by EVI1, we attempted to determine the essential promoter region of EGFR using luciferase assay. In the luciferase assays, we mainly used U87MG cells because these cells received few cell damages by the HilyMax compared to other GBM cell lines. The luciferase activity reduced significantly with pGL4-335 (59%) and pGL-265 (93%) than with pGL-377 (Fig. 3a). This result suggested that EGFR transcription was primarily driven by DNA derived from the 112 bp fragment, which was located − 377 bp to − 266 bp upstream of the EGFR promoter. When pGL4-377 was co-transfected with the EVI1 overexpression vector into five GBM cell lines (A172, LN229, U-87MG, U-251MG, and YKG-1), exogenous EVI1 enhanced the luciferase activities by 2.9-fold in YKG1 and by 11.4-fold in U-251MG (Fig. 3b). Thus, EVI1 had a positive influence on EGFR transcription in GBM cells. To determine the more important region (− 377 bp to − 336 bp vs. − 335 bp to − 266 bp) for the EVI1 regulation of EGFR transcription, luciferase activities were measured after co-transfection of the pGL4-265, pGL4-335, and pGL4-377 plasmids with the EVI1 overexpression vector (pMXs-EVI1) into the U-87MG cells. The luciferase activities were enhanced by exogenous EVI1 in both the pGL4-335 and pGL4-377 plasmids. Furthermore, the luciferase activity was significantly higher in pGL4-377 than in pGL4-335 (Fig. 3c). The results suggested that both EGFR promoter regions were important for the EVI1 regulation of EGFR transcription. We designated the promoter region in − 377 bp to − 266 bp as the EVI1 regulatory region (ERR). In ERR, − 377 bp to − 336 bp were designated as ERR1 and − 335 bp to − 266 bp were designated as ERR2 (Fig. 3d).
The role of the polypyrimidine stretches in the EVI1 regulatory region in the EVI1 activation of the EGFR promoter activity
We focused on the EVI1-binding sequences in ERR. EVI1 contains two independent DNA-binding domains, including the N-terminal and C-terminal domains. Previous reports showed that the EVI1 N-terminal domains recognized a GATA-like motif and the C-terminal domains recognized an ETS-like motif [3, 13]. Notably, ERR had no GATA-like motif, but it was rich in polypyrimidine stretches. We presumed that EVI1 needs at least two regions of ERR1 and ERR2 to regulate EGFR transcription. Furthermore, previous studies have shown that two regions of TC-rich direct-repeat sequences (general motif; TCCTCCTCC) in the EGFR promoter are essential for the regulation of EGFR transcription [14, 15]. Therefore, we generated eight EGFR mutant promoter constructs, viz., pGL4-377mt303, pGL4-377mt323, pGL4-377mt344, pGL4-377mt349, pGL4-377mt363, pGL4-377mt303/343, pGL4-377mt303/349, and pGL4-377mt303/363. We transfected these eight plasmids with the EVI1 overexpression vector into U-87 MG cells. As a result, the mutant plasmid had reduced EGFR transcriptional activity. In particular, transfection with pGL4-377mt303/343, pGL4-377mt303/349, and pGL4-377mt303/363 completely abolished EVI1 efficacy to enhance EGFR promoter activity (Fig. 3e).
The promoter activity of EGFR is enhanced by EVI1 via the C-terminal DNA-binding domain
The EVI1 mutant construct with deletion of the C-terminal DNA-binding domain (pCMV26-EVI1Δ8-10) was transfected with pGL4-377 into U-87MG, U-251MG, and YKG1 cells (Fig. 3f). EGFR promoter activity was enhanced in the presence of transfected wild-type EVI1 but markedly reduced after transfection of the EVI1 deletion mutant construct (pCMV26-EVI1Δ8-10). This suggested that the loss of the pCMV26-EVI1Δ8-10 transcriptional enhancer activity could be attributed to losing the capacity to bind to the EGFR promoter region.
EVI1 knockdown suppressed the proliferation of GBM cells
We investigated whether EVI1 knockdown affected the proliferation of A172 and YKG-1 cells. On day 3, the transfection of siRNAs against EVI1 significantly suppressed the proliferation of both cell types compared with the effects in the negative control (Fig. 4).
Discussion
In this study, we introduced the high tissue expression of EVI1 in patients with GBM might relate to poor prognosis. Next, we demonstrated that EVI1 regulated EGFR transcription and affected the proliferation of GBM cells. Further, we found that both TC-rich stretches in the EGFR promoter were essential for the EVI1 regulation of EGFR transcription. Only few reports on glioma focused on EVI1. In this study, we first reported that EVI1 regulated EGFR transcription in GBM cells.
EGFR amplification occurs in about 50% of primary GBM [16]. To date, several transcription factors have been reported to contribute to the regulation of EGFR transcription [14, 15, 17, 18]. Among these reports, the one by Johnson et al. is intriguing [14, 19] because they identified the two pairs of direct-repeat sequences, which conformed to the general motif TCCTCCTCC, on the EGFR promoter. We reported in this study that EVI1 also needed the TC-rich stretches in ERR, and the ability of EVI1 to regulate EGFR transcription depends on both ERR1 and ERR2 sites.
According to a previous study, the relations between EGFR expression and the prognosis of patients with GBM are controversial [20]. In this study, the result showed no significant relation between the EGFR-mRNA expression levels and the post-operative days (Supplementary Fig. 3). This result suggested that high-EVI1 expression could be correlated with worse prognosis of GBM depend on another mechanism unrelated to EGFR regulation.
According to the cBioPortal for cancer genomics data (https://www.cbioportal.org/index.do), the frequency of EVI1 genetic alteration was lower in glioma (2.1% of 283 cases) than in other solid cancers (lung squamous cell carcinoma, 44.1%; ovarian serous cyst adenoma, 33.8%; esophageal carcinoma, 25.5%; and neuroendocrine prostate cancer, 22.4%). Therefore, most researchers on glial tumors had less interest in this transcription factor. However, we showed that in patients with glioma, EVI1 might be related to poor prognosis.
There were some limitations to this report. First, our study population was relatively small and the design was retrospective. Second, our findings were only based on in vitro assays. To validate the role of EVI1 in GBM, in vivo assays are critical. Nevertheless, we showed the exciting potential of EVI1 to interrupt glioma cell proliferation. Therefore, further studies should clarify the role of EVI1 in patients with glioma.
Conclusion
We reported in this study that EVI1 regulated EGFR transcription and effect on the cell proliferation in GBM cells. The TC-rich stretches in the EGFR promoter (from − 377 to − 266 bp, relative to the EGFR translational start site) was essential for the EVI regulation of EGFR expression.
References
Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM, Miller TA, Pietryga DW, Scott WJ Jr, Potter SS (1991) A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell 65:677–689
Morishita K, Parker DS, Mucenski ML, Jenkins NA, Copeland NG, Ihle JN (1988) Retroviral activation of a novel gene encoding a zinc finger protein in IL-3-dependent myeloid leukemia cell lines. Cell 54:831–840
Funabiki T, Kreider BL, Ihle JN (1994) The carboxyl domain of zinc fingers of the Evi-1 myeloid transforming gene binds a consensus sequence of GAAGATGAG. Oncogene 9:1575–1581
Delwel R, Funabiki T, Kreider BL, Morishita K, Ihle JN (1993) Four of the seven zinc fingers of the Evi-1 myeloid-transforming gene are required for sequence-specific binding to GA(C/T)AAGA(T/C)AAGATAA. Mol Cell Biol 13:4291–4300
Li Y, Cheng CN, Verdun VA, Wingert RA (2014) Zebrafish nephrogenesis is regulated by interactions between retinoic acid, mecom, and Notch signaling. Dev Biol 386:111–122. https://doi.org/10.1016/j.ydbio.2013.11.021
Konantz M, Alghisi E, Muller JS, Lenard A, Esain V, Carroll KJ, Kanz L, North TE, Lengerke C (2016) Evi1 regulates Notch activation to induce zebrafish hematopoietic stem cell emergence. EMBO J. https://doi.org/10.15252/embj.201593454
Koos B, Bender S, Witt H, Mertsch S, Felsberg J, Beschorner R, Korshunov A, Riesmeier B, Pfister S, Paulus W, Hasselblatt M (2011) The transcription factor evi-1 is overexpressed, promotes proliferation, and is prognostically unfavorable in infratentorial ependymomas. Clin Cancer Res 17:3631–3637. https://doi.org/10.1158/1078-0432.CCR-11-0175
Hou A, Zhao L, Zhao F, Wang W, Niu J, Li B, Zhou Z, Zhu D (2016) Expression of MECOM is associated with unfavorable prognosis in glioblastoma multiforme. Onco Targets Ther 9:315–320. https://doi.org/10.2147/OTT.S95831
Bard-Chapeau EA, Jeyakani J, Kok CH, Muller J, Chua BQ, Gunaratne J, Batagov A, Jenjaroenpun P, Kuznetsov VA, Wei CL, D'Andrea RJ, Bourque G, Jenkins NA, Copeland NG (2012) Ecotopic viral integration site 1 (EVI1) regulates multiple cellular processes important for cancer and is a synergistic partner for FOS protein in invasive tumors. Proc Natl Acad Sci USA 109:2168–2173. https://doi.org/10.1073/pnas.1119229109
Purow BW, Sundaresan TK, Burdick MJ, Kefas BA, Comeau LD, Hawkinson MP, Su Q, Kotliarov Y, Lee J, Zhang W, Fine HA (2008) Notch-1 regulates transcription of the epidermal growth factor receptor through p53. Carcinogenesis 29:918–925. https://doi.org/10.1093/carcin/bgn079
Cenciarelli C, Marei HE, Zonfrillo M, Casalbore P, Felsani A, Giannetti S, Trevisi G, Althani A, Mangiola A (2017) The interference of Notch1 target Hes1 affects cell growth, differentiation and invasiveness of glioblastoma stem cells through modulation of multiple oncogenic targets. Oncotarget 8:17873–17886. https://doi.org/10.18632/oncotarget.15013
Jazaeri AA, Ferriss JS, Bryant JL, Dalton MS, Dutta A (2010) Evaluation of EVI1 and EVI1s (Delta324) as potential therapeutic targets in ovarian cancer. Gynecol Oncol 118:189–195. https://doi.org/10.1016/j.ygyno.2010.04.007
Yuasa H, Oike Y, Iwama A, Nishikata I, Sugiyama D, Perkins A, Mucenski ML, Suda T, Morishita K (2005) Oncogenic transcription factor Evi1 regulates hematopoietic stem cell proliferation through GATA-2 expression. EMBO J 24:1976–1987. https://doi.org/10.1038/sj.emboj.7600679
Johnson AC, Jinno Y, Merlino GT (1988) Modulation of epidermal growth factor receptor proto-oncogene transcription by a promoter site sensitive to S1 nuclease. Mol Cell Biol 8:4174–4184
Englert C, Hou X, Maheswaran S, Bennett P, Ngwu C, Re GG, Garvin AJ, Rosner MR, Haber DA (1995) WT1 suppresses synthesis of the epidermal growth factor receptor and induces apoptosis. EMBO J 14:4662–4675
Furnari FB, Cloughesy TF, Cavenee WK, Mischel PS (2015) Heterogeneity of epidermal growth factor receptor signalling networks in glioblastoma. Nat Rev Cancer 15:302–310. https://doi.org/10.1038/nrc3918
Ludes-Meyers JH, Subler MA, Shivakumar CV, Munoz RM, Jiang P, Bigger JE, Brown DR, Deb SP, Deb S (1996) Transcriptional activation of the human epidermal growth factor receptor promoter by human p53. Mol Cell Biol 16:6009–6019
Vaughan CA, Pearsall I, Singh S, Windle B, Deb SP, Grossman SR, Yeudall WA, Deb S (2016) Addiction of lung cancer cells to GOF p53 is promoted by up-regulation of epidermal growth factor receptor through multiple contacts with p53 transactivation domain and promoter. Oncotarget 7:12426–12446. https://doi.org/10.18632/oncotarget.6998
Johnson AC (1996) Activation of epidermal growth factor receptor gene transcription by phorbol 12-myristate 13-acetate is mediated by activator protein 2. J Biol Chem 271:3033–3038
Ohgaki H, Dessen P, Jourde B, Horstmann S, Nishikawa T, Di Patre PL, Burkhard C, Schuler D, Probst-Hensch NM, Maiorka PC, Baeza N, Pisani P, Yonekawa Y, Yasargil MG, Lutolf UM, Kleihues P (2004) Genetic pathways to glioblastoma: a population-based study. Cancer Res 64:6892–6899. https://doi.org/10.1158/0008-5472.CAN-04-1337
Acknowledgements
We would like to thank Dr. Kazuko Kaneda, Dr. Shingo Nakahata, Dr. Sohei Mizuguchi, and Ms. Ayumi Nagatomo for their assistance. We would like to thank Enago (www.enago.jp) for the English language review. This study was supported by a Grant-in-Aid for Clinical Research from Miyazaki University Hospital, JSPS KAKENHI Grant Number 25462275, 19K09486, and MSD [Affiliated company of Merck & Co., Inc., Whitehouse Station, NJ, USA].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
This study was approved by the Ethics Committee of the University of Miyazaki Hospital, Miyazaki, Japan,
Informed consent
All patients and their families gave informed consent for the use of the resected tissues.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Mizuguchi, A., Yamashita, S., Yokogami, K. et al. Ecotropic viral integration site 1 regulates EGFR transcription in glioblastoma cells. J Neurooncol 145, 223–231 (2019). https://doi.org/10.1007/s11060-019-03310-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-019-03310-z