Abstract
Purpose
The TruSight Tumor 170 (TST-170) panel consists of a DNA workflow for the identification of single-nucleotide variants, small insertions and deletions, and copy number variation, as well as a panel of 55 genes for a RNA workflow for the identification of splice variants and gene fusions. To date, the application of TST-170 in diffuse gliomas (DGs) has not been described.
Methods
We analyzed 135 samples of DG, which were diagnosed by WHO criteria based on histological features and conventional molecular tests including immunostaining, 1p/19q FISH, and analysis of MGMT methylation and TERT promoter mutation.
Results
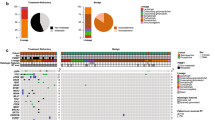
A total of 135 cases consisted of 38 IDH-mutant [17 astrocytoma (AC), 13 oligodendroglioma (OD) and eight glioblastoma (GBM)], 87 IDH-wildtype (six AC, three OD and 78 GBM), and 10 diffuse midline glioma, H3K27M-mutant. DNA analysis enabled the detection of all mutations identified in these samples by conventional techniques, and the results were highly comparable to the known mutations in each subtype. RNA analysis detected four fusion genes including PTPRZ1–MET, FGFR3–TACC3, FAM131B–BRAF, and RET–CCDC6 and one splicing variant (EGFR vIII mutant). Clustered copy number loss in 1p and 19q loci genes were detected in 1p/19q-codeleted OD.
Conclusions
The application of TST-170 panel based NGS in clinical and laboratory setting is expected to improve diagnostic accuracy and prognostication. Most benefits are expected in IDH-wildtype DG, a group of genetically heterogenous tumors harboring DNA sequence changes, copy number alterations, and fusions in a large number of oncogenes and tumor suppressor genes.


Similar content being viewed by others
References
Goodenberger ML, Jenkins RB (2012) Genetics of adult glioma. Cancer Genet 205(12):613–621. https://doi.org/10.1016/j.cancergen.2012.10.009
Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW (2016) The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131(6):803–820. https://doi.org/10.1007/s00401-016-1545-1
Huse JT, Aldape KD (2014) The evolving role of molecular markers in the diagnosis and management of diffuse glioma. Clin Cancer Res 20(22):5601–5611. https://doi.org/10.1158/1078-0432.CCR-14-0831
Nikiforova MN, Wald AI, Roy S, Durso MB, Nikiforov YE (2013) Targeted next-generation sequencing panel (ThyroSeq) for detection of mutations in thyroid cancer. J Clin Endocrinol Metab 98(11):E1852–E1860. https://doi.org/10.1210/jc.2013-2292
Nikiforova MN, Wald AI, Melan MA, Roy S, Zhong S, Hamilton RL, Lieberman FS, Drappatz J, Amankulor NM, Pollack IF, Nikiforov YE, Horbinski C (2016) Targeted next-generation sequencing panel (GlioSeq) provides comprehensive genetic profiling of central nervous system tumors. Neuro Oncol 18(3):379–387. https://doi.org/10.1093/neuonc/nov289
Fisher KE, Zhang L, Wang J, Smith GH, Newman S, Schneider TM, Pillai RN, Kudchadkar RR, Owonikoko TK, Ramalingam SS, Lawson DH, Delman KA, El-Rayes BF, Wilson MM, Sullivan HC, Morrison AS, Balci S, Adsay NV, Gal AA, Sica GL, Saxe DF, Mann KP, Hill CE, Khuri FR, Rossi MR (2016) Clinical validation and implementation of a targeted next-generation sequencing assay to detect somatic variants in non-small cell lung, melanoma, and gastrointestinal malignancies. J Mol Diagn 18(2):299–315. https://doi.org/10.1016/j.jmoldx.2015.11.006
Louis DN, Wesseling P, Paulus W, Giannini C, Batchelor TT, Cairncross JG, Capper D, Figarella-Branger D, Lopes MB, Wick W, van den Bent M (2018) cIMPACT-NOW update 1: not otherwise specified (NOS) and not elsewhere classified (NEC). Acta Neuropathol 135(3):481–484. https://doi.org/10.1007/s00401-018-1808-0
Louis DN, Giannini C, Capper D, Paulus W, Figarella-Branger D, Lopes MB, Batchelor TT, Cairncross JG, van den Bent M, Wick W, Wesseling P (2018) cIMPACT-NOW update 2: diagnostic clarifications for diffuse midline glioma, H3 K27M-mutant and diffuse astrocytoma/anaplastic astrocytoma, IDH-mutant. Acta Neuropathol 135(4):639–642. https://doi.org/10.1007/s00401-018-1826-y
Brat DJ, Aldape K, Colman H, Holland EC, Louis DN, Jenkins RB, Kleinschmidt-DeMasters BK, Perry A, Reifenberger G, Stupp R, von Deimling A, Weller M (2018) cIMPACT-NOW update 3: recommended diagnostic criteria for “diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV”. Acta Neuropathol 136(5):805–810. https://doi.org/10.1007/s00401-018-1913-0
Bechet D, Gielen GG, Korshunov A, Pfister SM, Rousso C, Faury D, Fiset PO, Benlimane N, Lewis PW, Lu C, David Allis C, Kieran MW, Ligon KL, Pietsch T, Ellezam B, Albrecht S, Jabado N (2014) Specific detection of methionine 27 mutation in histone 3 variants (H3K27M) in fixed tissue from high-grade astrocytomas. Acta Neuropathol 128(5):733–741. https://doi.org/10.1007/s00401-014-1337-4
Reuss DE, Sahm F, Schrimpf D, Wiestler B, Capper D, Koelsche C, Schweizer L, Korshunov A, Jones DT, Hovestadt V, Mittelbronn M, Schittenhelm J, Herold-Mende C, Unterberg A, Platten M, Weller M, Wick W, Pfister SM, von Deimling A (2015) ATRX and IDH1-R132H immunohistochemistry with subsequent copy number analysis and IDH sequencing as a basis for an “integrated” diagnostic approach for adult astrocytoma, oligodendroglioma and glioblastoma. Acta Neuropathol 129(1):133–146. https://doi.org/10.1007/s00401-014-1370-3
Ritterhouse LL, Barletta JA (2015) BRAF V600E mutation-specific antibody: a review. Semin Diagn Pathol 32(5):400–408. https://doi.org/10.1053/j.semdp.2015.02.010
Takami H, Yoshida A, Fukushima S, Arita H, Matsushita Y, Nakamura T, Ohno M, Miyakita Y, Shibui S, Narita Y, Ichimura K (2015) Revisiting TP53 mutations and immunohistochemistry—a comparative study in 157 diffuse gliomas. Brain Pathol 25(3):256–265. https://doi.org/10.1111/bpa.12173
Woehrer A, Sander P, Haberler C, Kern S, Maier H, Preusser M, Hartmann C, Kros JM, Hainfellner JA, Research Committee of the European Confederation of Neuropathological Societies (2011) FISH-based detection of 1p 19q codeletion in oligodendroglial tumors: procedures and protocols for neuropathological practice—a publication under the auspices of the Research Committee of the European Confederation of Neuropathological Societies (Euro-CNS). Clin Neuropathol 30(2):47–55
McNamara MG, Jiang H, Lim-Fat MJ, Sahebjam S, Kiehl TR, Karamchandani J, Coire C, Chung C, Millar BA, Laperriere N, Mason WP (2017) Treatment outcomes in 1p19q Co-deleted/partially deleted gliomas. Can J Neurol Sci 44(3):288–294. https://doi.org/10.1017/cjn.2016.420
Carter JH, McNulty SN, Cimino PJ, Cottrell CE, Heusel JW, Vigh-Conrad KA, Duncavage EJ (2017) Targeted next-generation sequencing in molecular subtyping of lower-grade diffuse gliomas: application of the World Health Organization’s 2016 revised criteria for central nervous system tumors. J Mol Diagn 19(2):328–337. https://doi.org/10.1016/j.jmoldx.2016.10.010
Kaneshiro D, Kobayashi T, Chao ST, Suh J, Prayson RA (2009) Chromosome 1p and 19q deletions in glioblastoma multiforme. Appl Immunohistochem Mol Morphol 17(6):512–516. https://doi.org/10.1097/PAI.0b013e3181a2c6a4
Mukasa A, Takayanagi S, Saito K, Shibahara J, Tabei Y, Furuya K, Ide T, Narita Y, Nishikawa R, Ueki K, Saito N (2012) Significance of IDH mutations varies with tumor histology, grade, and genetics in Japanese glioma patients. Cancer Sci 103(3):587–592. https://doi.org/10.1111/j.1349-7006.2011.02175.x
Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A, Felsberg J, Wolter M, Mawrin C, Wick W, Weller M, Herold-Mende C, Unterberg A, Jeuken JW, Wesseling P, Reifenberger G, von Deimling A (2009) Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol 118(4):469–474. https://doi.org/10.1007/s00401-009-0561-9
Padul V, Epari S, Moiyadi A, Shetty P, Shirsat NV (2015) ETV/Pea3 family transcription factor-encoding genes are overexpressed in CIC-mutant oligodendrogliomas. Genes Chromosomes Cancer 54(12):725–733. https://doi.org/10.1002/gcc.22283
Nauen D, Haley L, Lin MT, Perry A, Giannini C, Burger PC, Rodriguez FJ (2016) Molecular analysis of pediatric oligodendrogliomas highlights genetic differences with adult counterparts and other pediatric gliomas. Brain Pathol 26(2):206–214. https://doi.org/10.1111/bpa.12291
Suri V, Jha P, Agarwal S, Pathak P, Sharma MC, Sharma V, Shukla S, Somasundaram K, Mahapatra AK, Kale SS, Sarkar C (2011) Molecular profile of oligodendrogliomas in young patients. Neuro Oncol 13(10):1099–1106. https://doi.org/10.1093/neuonc/nor146
Zhang J, Wu G, Miller CP, Tatevossian RG, Dalton JD, Tang B, Orisme W, Punchihewa C, Parker M, Qaddoumi I, Boop FA, Lu C, Kandoth C, Ding L, Lee R, Huether R, Chen X, Hedlund E, Nagahawatte P, Rusch M, Boggs K, Cheng J, Becksfort J, Ma J, Song G, Li Y, Wei L, Wang J, Shurtleff S, Easton J, Zhao D, Fulton RS, Fulton LL, Dooling DJ, Vadodaria B, Mulder HL, Tang C, Ochoa K, Mullighan CG, Gajjar A, Kriwacki R, Sheer D, Gilbertson RJ, Mardis ER, Wilson RK, Downing JR, Baker SJ, Ellison DW, St. Jude Children’s Research Hospital-Washington University Pediatric Cancer Genome (2013) Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet 45(6):602–612. https://doi.org/10.1038/ng.2611
Cancer Genome Atlas Research Network (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455(7216):1061–1068. https://doi.org/10.1038/nature07385
Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, Stegh AH, Bradner JE, Ligon KL, Brennan C, Chin L, DePinho RA (2007) Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science 318(5848):287–290. https://doi.org/10.1126/science.1142946
Bao ZS, Chen HM, Yang MY, Zhang CB, Yu K, Ye WL, Hu BQ, Yan W, Zhang W, Akers J, Ramakrishnan V, Li J, Carter B, Liu YW, Hu HM, Wang Z, Li MY, Yao K, Qiu XG, Kang CS, You YP, Fan XL, Song WS, Li RQ, Su XD, Chen CC, Jiang T (2014) RNA-seq of 272 gliomas revealed a novel, recurrent PTPRZ1-MET fusion transcript in secondary glioblastomas. Genome Res 24(11):1765–1773. https://doi.org/10.1101/gr.165126.113
Di Stefano AL, Fucci A, Frattini V, Labussiere M, Mokhtari K, Zoppoli P, Marie Y, Bruno A, Boisselier B, Giry M, Savatovsky J, Touat M, Belaid H, Kamoun A, Idbaih A, Houillier C, Luo FR, Soria JC, Tabernero J, Eoli M, Paterra R, Yip S, Petrecca K, Chan JA, Finocchiaro G, Lasorella A, Sanson M, Iavarone A (2015) Detection, characterization, and inhibition of FGFR-TACC fusions in IDH wild-type glioma. Clin Cancer Res 21(14):3307–3317. https://doi.org/10.1158/1078-0432.CCR-14-2199
Bayliss J, Mukherjee P, Lu C, Jain SU, Chung C, Martinez D, Sabari B, Margol AS, Panwalkar P, Parolia A, Pekmezci M, McEachin RC, Cieslik M, Tamrazi B, Garcia BA, La Rocca G, Santi M, Lewis PW, Hawkins C, Melnick A, David Allis C, Thompson CB, Chinnaiyan AM, Judkins AR, Venneti S (2016) Lowered H3K27me3 and DNA hypomethylation define poorly prognostic pediatric posterior fossa ependymomas. Sci Transl Med 8(366):366ra161. https://doi.org/10.1126/scitranslmed.aah6904
Wang L, Li Z, Zhang M, Piao Y, Chen L, Liang H, Wei Y, Hu Z, Zhao L, Teng L, Lu D (2018) H3 K27M-mutant diffuse midline gliomas in different anatomical locations. Hum Pathol 78:89–96. https://doi.org/10.1016/j.humpath.2018.04.015
Solomon DA, Wood MD, Tihan T, Bollen AW, Gupta N, Phillips JJ, Perry A (2016) Diffuse midline gliomas with histone H3-K27M mutation: a series of 47 cases assessing the spectrum of morphologic variation and associated genetic alterations. Brain Pathol 26(5):569–580. https://doi.org/10.1111/bpa.12336
Meyronet D, Esteban-Mader M, Bonnet C, Joly MO, Uro-Coste E, Amiel-Benouaich A, Forest F, Rousselot-Denis C, Burel-Vandenbos F, Bourg V, Guyotat J, Fenouil T, Jouvet A, Honnorat J, Ducray F (2017) Characteristics of H3 K27M-mutant gliomas in adults. Neuro Oncol 19(8):1127–1134. https://doi.org/10.1093/neuonc/now274
Funding
This work was supported by the Brain Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Science, ICT & Future Planning (Grant No. 2016M3C7A1913844).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Na, K., Kim, HS., Shim, H.S. et al. Targeted next-generation sequencing panel (TruSight Tumor 170) in diffuse glioma: a single institutional experience of 135 cases. J Neurooncol 142, 445–454 (2019). https://doi.org/10.1007/s11060-019-03114-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-019-03114-1