Abstract
The Apoptosis is an intensely studied mechanism for cancer therapy. Various genes, drugs, and molecules have been combined to potentiate the apoptotic effect against tumor cells. Magnetic nanocomplexes with biopolymers have been proposed as nanocarriers for delivery of active molecules and improve apoptosis induction. To these purpose were formulated magnetic nanoparticles with curcumin coated chitosan (MNPs-CHI-TPP-CUR) and magnetic nanoparticles with pCEM-TRAIL plasmid coated chitosan (MNPs-CHI-TPP-TRAIL) for use alone or in combination to induce the apoptosis of B16F10 tumor cells because curcumin is capable of overexpressing receptors TRAIL required for apoptosis induction. Also, curcumin (CUR) has been used as a photosensitizer in photodynamic therapy (PDT). In this work, it was produced and characterized nanoparticles with curcumin and nanoparticles with TRAIL gene. Both types of nanoparticles were ~ 200 nm size. The results showed that the combination of (MNPs-CHI-TPP-CUR) and (MNPs-CHI-TPP-TRAIL) induced a significant increase in cellular death within 48 h compared with the effect of each nanocomplex individually, demonstrating a synergic effect. This is the first description the synergic interaction between TRAIL gene and curcumin drug to improve the apoptosis in cancer cells. Also, it was determined that cellular death was potentiated when the treated cells were exposed to laser photoactivation with wavelengths of 405 nm and 532 nm for 15 min.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cancer is one of the diseases with the highest incidence and mortality rate in the world. Conventional treatments are chemotherapies or radiotherapies that in turn cause toxicity and adverse effects, which leads to the search for new specific therapies to be developed to attack cancer cells without affecting healthy adjacent tissues or organs [1].
The use of nanoparticles as directed drug delivery systems is becoming more widespread and promising as it allows the encapsulation of chemotherapeutic agents. Chitosan nanoparticles with the cross-linking agent as tripolyphosphate of pentasodium (TPP) have already been used to ship drugs to tumors [2, 3]. One of the candidate drugs with anticancer activity and that blocks crucial steps of metastasis by altering the molecules involved in metastatic tumor progression is curcumin, its efficacy for biological applications has long been studied, and its versatile properties have allowed researchers for drug delivery applications to make it reach the desired site in an appropriate carrier such as magnetic nanoparticles [4]. Animal and human studies have already been conducted showing that curcumin is safe and well tolerated, even when doses are administered later. In addition, the effects of curcumin can be enhanced with the application of light in therapeutic mode; this is known as photodynamic therapy which uses an association of a photosensitizing agent with light at an appropriate wavelength [5,6,7]. The interaction of the agent with light in the presence of oxygen causes reactive species that promote cell damage and death [8]. The use of curcumin has not yet been tested in clinical trials there are many in vitro studies that show the effect of CUR on PDT for cancer cells [9, 10].
One of the strategies to attack cancer is the combination of therapies that have proven to be more successful than monotherapy in reducing toxicity and synergistic effects. TRAIL is an example of a molecule that selectively kills malignant cells, but not normal. Also has been shown to have synergistic activity with curcumin [11,12,13,14,15].
Until today, the synergistic effect between TRAIL and curcumin has only been demonstrated at the level of the molecules, either free or in the form of nanoparticles [16]. Therefore, the purpose of the current study was to synthesize and characterize magnetic nanoparticles of chitosan with TRAIL and CUR (MNPs-CHI-TPP-TRAIL) and (MNPs-CHI-TPP-CUR) to evaluate cellular death effect and photodynamic effects at different wavelengths (405 and 532 nm) in a melanoma cell line. This as a possibility of using nanoformulations as a chemogenephotodynamic therapy strategy [17].
Materials and Methods
Materials
Chitosan (25 kDa) was purchased from CoyoteFoods Biopolymer and Biotechnology Mexico. Curcumin was obtained from Merck Millipore®, and cell culture media, fetal bovine serum, and cell culture supplements were obtained from Thermo Fisher Scientific (Waltham, MA, USA). Tripolyphosphate (TPP) was purchased from Sigma-Aldrich (St Louis, MO, USA). MNPs were obtained from OZ Biosciences (CombiMag; Marseille, France). Magnetic fluorescent nanoparticles (MNPs) composed of an iron oxide (SPIO) core and covered with a biocompatible silica layer and an organic fluorophore with a spectrum similar to the cyanine fluorochrome 5.5 (MaxViagenTM -710 nm, NVIGEN) covalently attached. A plasmid purification kit was purchased from Thermo Fisher Scientific. Magnetofector plates (A 8 × 12 cm) with 96 wells were obtained from Boca Scientific (Dedham, MA, USA) for use with a green and red laser (ZH-303) of a 532–650 nm wavelength range.
Plasmid TRAIL
pCEM-TRAIL was constructed and characterized in our laboratory. It encodes the TRAIL gene and contains the promoter sequence (273 bp) of the 70-kDa human heat shock protein (HSP70). The expression of the gene controlled by this promoter is induced when exposed to a magnetic field (was constructed and characterized previously) [18].
Melanoma cell line
The B16F10 mouse melanoma cell line from our laboratory cell bank was used. The cells were maintained at 37 °C in 5% CO2.
Production of the magnetic nanoparticles with chitosan and the TRAIL plasmid: MNPs-CHI-TPP-TRAIL
For the synthesis of nanoparticles, the ionic gelation method was performed according to the protocol of Calvo et al. (1997) [19] with the charges of chitosan and TPP used as crossover agents. Chitosan solution (2 mg/mL at pH 5.5), 50 mL of TPP solution, and pCEM-TRAIL (0.5 to 1 μg/µL) were prepared. The fluorescent magnetic nanoparticle (MNP) solution was prepared from a stock solution of 1 μg/ml in sterile PBS diluted 1:10. The TPP, pDNA, and curcumin solutions were added dropwise to the chitosan solution to the same volume while mixing at 950 rpm/min at room temperature. The suspension was gently shaken for 1 h at room temperature.
To calculate the amount of chitosan required for each nanocomplex, the following formula was used according to the proportional CHI:pDNA ratio:
TPP to chitosan was added at a 1:2 ratio, and the amount of pDNA was 1 μg. Briefly, MNPs, pCEM-TRAIL, and TPP were mixed in a tube and, in another tube, were mixed chitosan and TPP. Then both were vortex for 90 s.
The DNA-binding ability of chitosan was evaluated by agarose gel electrophoresis. Complexes containing 1 µg of DNA and different proportions of chitosan were loaded into individual wells of a 1% agarose gel, electrophoresed at 80 V for 40 min, and stained with ethidium bromide.
Production of magnetic chitosan nanoparticles with curcumin: MNPs-CHI-TPP-CUR
To standardize the production of the MNPs-CHI-TPP-CUR, the ionic gelation procedure based on Calvo et al. (1997) [19] was used with procedures previously performed by our laboratory. The construction of these nanocomplexes was similar to that of MNPs-CHI-TPP-TRAIL: the reagents were placed in two tubes: chitosan (and curcumin with or without Tween 80, according to the test) and the MNPs and µL of TPP were placed in another tube and vortex and agitated at 950 rpm at room temperature for 1 h. A large-scale ionic gelling protocol based on the synthesis described and standardized. Nanoparticle synthesis was performed in a beaker with magnetic stirring. A total of 750 µL of chitosan (2 mg/mL at pH 5.5) and 25 µL of curcumin (300 µg/mL dissolved in absolute ethanol) were added to the beaker and stirred for 30 min at a speed of 990 rpm (Corning™ PC-220 Hot Plate/Stirrers) in the dark.
Characterization of the nanocomplexes
Characterization of the MNPs-CHI-TPP-CUR by transmission electron microscopy (TEM)
For the observations, one drop of the aqueous dispersion of nanoparticles was added to a coated copper grid, air-dried in vacuum desiccators, and then examined under an electron microscope.
Size and homogeneity
The size and homogeneity of the nanocomplexes were determined by dynamic light dispersion using a Zetasizer apparatus (ZS90, Malvern, UK). The nanocomplexes were calibrated to 1000 μL with filtered distilled water and loaded into 1-mL polystyrene cells.
The zeta potential was obtained by electrophoretic light dispersion using a Zetasizer device (ZS90, Malvern). The nanocomplexes were calibrated to 1 ml with filtered sterile distilled water and then filled with an insulin syringe in a folded capillary cell (DTS1070, Malvern, UK).
FTIR analysis
The FTIR spectra of magnetic nanoparticles (MNPs), curcumin (CUR), chitosan (CHI), and magnetic nanoparticles of chitosan and curcumin (MNPs-CHI-TPP-CUR) were obtained using PerkinElmer spectrum 400 FTIR spectrometer over the range 4000–400 cm−1.
UV–visible spectroscopy analysis
An ultraviolet–visible near-infrared spectrophotometer Varioskan LUX was used to determine whether curcumin was conjugated to the MNPs-CHI-TPP-CUR.
MNPs-CHI-TPP-TRAIL plasmid release
The nanocomplexes were constructed with the aforementioned specifications. When the time required for ionic gelation elapsed, the nanocomplexes were centrifuged at 14,800 rpm for 30 min in a microcentrifuge (Sorvall Legend Micro 21R, Thermo Scientific), the supernatant was collected, and the pDNA was quantified by spectrophotometry (NanoDrop 2000, Thermo Scientific). The nanocomplex pellet was resuspended with the addition of PBS by pipetting, and the nanocomplexes were incubated again with agitation (950 rpm). This procedure was repeated every hour for 6 h and then performed at 12, 24, and 48 h. The pDNA obtained in the first supernatant collected corresponded to pDNA and was not retained; therefore, it was possible to calculate the percentage of pDNA released.
Curcumin release from nanocomplexes
After the hour required for ionic gelation, samples were centrifuged at 14,800 rpm for 30 min in a microcentrifuge (Sorvall Legend Micro 21R, Thermo Scientific), and the supernatant was collected and deposited on a 96-well plate (100 µL per well) until the supernatant was depleted. Subsequently, the curcumin absorbance reading was performed using a Varioskan Lux (Thermo Fisher Scientific) plate reader at a wavelength of 420 nm. To quantify curcumin based on absorbance, linear regression was performed on the basis of known curcumin concentrations.
Nanocomplexes-induced cytotoxicity in B16F10 cells assessed by MTT viability test
B16F10 cells (3 × 103) were seeded at 100 µL in Dulbecco’s modified Eagle’s medium supplemented with bovine fetal serum and 1% antibiotic–antimycotic and incubated at 37 °C with 5% CO2 for 24 h before treatment until the cells were 80% confluent.
Once the B16F10 cells reached the desired confluency, the following treatments were administered: control, vehicle, nanocomplexes with pCEM-TRAIL, nanocomplexes with curcumin, and dual nanocomplexes (MNPs-CHI-TPP-TRAIL and MNPs-CHI-TPP-CUR). The experiments were performed in triplicate on the same plate with three replicates to evaluate the effect after 24, 48, and 72 h. At the start of the cell incubation at 37 °C with 5% CO2, each plate was exposed to a magnetic field for a period of 15 min. The absorbance was measured at 540 nm using a plate reader (Synergy 2, BioTek), and the viability results were plotted according to the treatment using the appropriate control reference to determine the percentage of viable cells.
Evaluation of B16F10 cell apoptosis exposed to different nanocomplexes as assessed by Annexin
In this experiment, we determined the effect of MNPs-CHI-TPP with CUR and TRAIL in B16F10 cells. With the combination of both nanoparticles, the effect was more notable, indicating synergism in the induction of apoptosis of B16F10 cells.
The transfected cells will be analyzed 24 h after transfection with the apoptosis detection kit by annexin V-FITC (Calbiochem®), which is based on the detection of phosphatidyl serine molecules in the outer cell membrane, an indication of early apoptosis, through the use of annexin V. After 24 h after the transfection, the cells were taken off. Then 10 μL of binding reagent was added followed by 1.25 μL of V-FITC annex solution. Samples were incubated for 15 min at room temperature (10–24 °C) in dark conditions. Once the incubation time has elapsed, the samples are centrifuged 1′000 g for 5 min at room temperature, and the medium is removed. The cells were resuspended in 0.5 mL of binding buffer, and 10 μL of propidium iodide was added. The samples were finally taken to the inverted fluorescence microscope (VWR®).
Nanocomplexes-induced cytotoxicity in B16F10 cells exposed to wavelengths of 405 nm and 532 nm as assessed by MTT viability test
B16F10 cells (3 × 103) were seeded and incubated at 37 °C with 5% CO2 for 24 h before treatment until the cells were 80% confluent, at which time the following treatments were administered: control, nanocomplexes with pCEM-TRAIL, nanocomplexes with curcumin, and dual treatment (MNPs-CHI-TPP-TRAIL and MNPs-CHI-TPP-CUR). Treatments were performed in triplicate on the same plate to evaluate the effect after 24 h of incubation at 37 °C and 5% CO2, and each plate was exposed to a magnetic field for a period of 15 min.
After the first 3 h of incubation, the plates were separated into two groups to irradiate them at 405 nm (100 mW) or 532 nm (100 mW) (ZH 303) for 5 min. Subsequently, the cells were placed in the incubator. Once the incubation time (24 h) had elapsed, and the determination of cellular viability was carried out. The absorbance at 540 nm was measured using a plate reader (Synergy 2, BioTek), and the viability results were plotted according to the treatment, using the appropriate control reference to determine the percentage of viable cells.
Cell cycle analysis by flow cytometry
Cell cycle evaluation was performed, using propidium iodide (PI) staining to intracellular DNA quantification by flow cytometry, 1 × 106 B16F10 cells were seeded in 6-well dishes, and exposed to the CC50 dose of MNPs-CHI-TPP-TRAIL and MNPs-CHI-TPP-CUR and incubated for 24 h. Later, cells were collected, washed with PBS, and fixed with 70% ethanol overnight at − 20 °C. The fixed cells were washed twice in PBS and then incubated with 50 μL of RNase solution (10 μg/mL in sterile PBS) for 30 min, and then 5μL of propidium iodide (1 mg/mL in sterile PBS) were added and incubated at room temperature in the dark for 15 min. Flow cytometric analysis was performed by a BD AccuriC6 flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). For each analysis, 100,000 events were collected and analyzed using FlowJo software (LLC, Ashland, Oregon USA).
Analysis of data
Data were presented as mean ± SD. The significant differences between means analyzed with two-way ANOVAs with Tukey’s test were performed for multiple comparisons of the means. The tests and the data-based graph generation were carried out with GraphPad version 8.2 statistical software.
Results
Image of magnetic fluorescent nanoparticles (MNPs)
The morphology of magnetic fluorescent nanoparticles naked (Fig. 1) showed a small size on the nanoscale with approximately 20 nm.
Characterization of MNPs-CHI-TPP-TRAIL
The evaluated ratios of pCEM-TRAIL:chitosan were 4:1, 8:1, 12:1, 16:1, 20:1, 24:1, and 28:1., and as a control sample, 1 µg of pDNA was used (Fig. 2).
Size, homogeneity, and potential zeta (ζ) characterization of MNPs-CHI-TPP-TRAIL
The optimal formulation for the synthesis of nanocomplexes was standardized by varying the CHI:TPP ratio, final volume, solvent dilution, and sonication parameters before and after ionic gelation. Figure 3A shows the graph obtained for the size assessment of the nanocomplexes based on the standardized formulation. An average sample size of ~ 206.2 nm was observed with an intensity of 100%, a polydispersity index of 0.192, and good quality. In addition, the zeta potential of the nanocomplexes is shown in Fig. 3B. The ζ potential is positive with an average of ~ 19.6 mV in 100% of the sample.
A Size and homogeneity of MNPs-CHI-TPP-TRAIL nanocomplexes. The nanocomplexes have an average size (Z-average) of 206.2 and a PdI (polydispersity index) of 0.192 and good quality. B Z potential of the MNPs-CHI-TPP-TRAIL nanocomplexes. The nanocomplexes have a zeta potential (mV) of + 19.5 in addition to good quality
Characterization of the pDNA release profile of the MNPs-CHI-TPP-TRAIL nanocomplexes
The nanocomplexes were incubated for 24 h to quantify the plasmidic DNA. Retention of ~ 80% was observed 30 min after the synthesis of the nanocomplexes (Fig. 4).
In vitro drug release of pDNA from chitosan nanocomplexes formulated with TPP and without TPP. The concentration of pDNA was detected by absorbance at different times, including 0.5, 2, 3.5, 5, 6.5, 12, and 24.5 h. The cumulative release profiles of pDNA from nanocomplexes exhibited similar kinetics of sustained release. Specifically, the total cumulative release of pDNA from nanocomplexes without TPP was 78.6% within 24.5 h, compared to a total cumulative release of 100% of the pDNA from nanocomplexes with TPP. The points represent the mean ± SD from a triplicate experiment
Assessment of curcumin cytotoxicity in B16F10 cells by feasibility test with an MTT-forming reaction
Figure 5 shows the percentage of viable B16F10 cells at different curcumin concentrations (0.0679–43.4334 µM) in medium (DMEM + 10% SFT + 1% antibiotic) for different exposure times (24, 48, and 72 h). The LD50 was observed at a concentration of 4,0719 µM for the 24- and 48-h exposure and at 1,018 µM for the 72-h exposure.
Characterization of the homogeneity and size of the MNPs-CHI-TPP-CUR nanocomplexes. Shape and size characterization of the MNPs-CHI-TPP-CUR nanocomplexes as assessed by TEM composition and FTIR
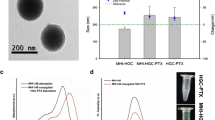
The magnetic nanoparticles with chitosan and curcumin (MNPs-CHI-TPP-CUR) analyzed by transmission electron microscopy showed a spherical shape and well-dispersed sample (Fig. 6A). For the optimal formulation, 2 mg/mL chitosan was added at pH 5.5, 0.86 mg/mL TPP was added at pH 3.0, the CHI:TPP ratio (v:v) was 2:1, the CHI:CUR ratio (v:v) was 30, and 25 µL of 300 µg/mL curcumin dissolved in absolute ethanol was added without Tween 80. Nanocomplexes with the standardized formulation showed an average sample size of ~ 258.6 nm with an intensity of 100%, a polydispersity index of 0.110, and good quality (Fig. 6B). In the FTIR spectrum of curcumin, the highest frequency bands observed within 2700–3000 cm−1 region are assigned to the aromatic C–H stretches. MNPs show a prominent peak of Fe–O vibrations at approximately 576 cm−1 which corresponds to the vibration of the Fe–O bonds in the crystalline lattice of Fe3O4 [20]. Therefore, there was no change in the chemical composition of the samples before and after the formation of nanoparticles process (Fig. 6C).
A Analysis of the nanocomplexes by scanning electron microscopy (SEM). B Size and homogenity of The MNPs-CHI-TPP-CUR. Average size (Z-average) of 258.6 nm and a PdI (polydispersity index) of 0.110 and good quality. C FTIR spectra of magnetic nanoparticles (MNPs-CUR), curcumin (CUR), and magnetic nanoparticles of curcumin (MNPs). D Ultraviolet spectra of magnetic nanoparticles (MNPs), magnetic nanoparticles of chitosan and curcumin (MNPsCHI-TPP-CUR), curcumin (CUR), and chitosan (CHI)
The ultraviolet spectra for MNPs, CHI, CUR, and MNPs-CHI-TPP-CUR are shown in Fig. 6D. An absorption maximum for MNPs is 285 nm, CHI206 nm, CUR 415 nm, and the absorption peak of MNPs-CHI-TPP-CUR 270 nm clearly indicated successful contact between the components.
Determination of curcumin encapsulation and release in MNPs-CHI-TPP-CUR nanocomplexes
Figure 7 corresponds to the calibration curve and slope production as well as the curcumin release assay, which was based on the percentage of curcumin retained in the chitosan-based magnetic nanocomplexes with curcumin. A retention of 55% of total curcumin used in the synthesis of the nanocomplexes was observed. Curcumin was progressively released until reaching a high level of release at 12 h and almost total release at 24 h.
Cytotoxicity in B16F10 cells induced by nanocomplexes and treatments assessed by MTT
Images of confocal microscopy of B16F10 cells transfected with different nanoformulations (Fig. 8A) and the viability at different times (24, 48, and 72 h) are shown in Fig. 8B.
A Confocal microscopy images of B16F10 morphology exposed to nanocomplexes at different times (24, 48, and 72 h). B Viability assay of B16F10 cells exposed to different formulations of nanoparticles (MNPs-CHI-TPP-TRAIL, MNPs-CHI-TPP-CUR, or both) after 24, 48, and 72 h. Statistical analyses were performed by second-way ANOVA followed by Tukey’s multiple comparison test. Symbols indicate significance *p < 0.05
Evaluation of early apoptosis and late apoptosis in B16F10 cells with different nanoformulations of MNPs with annexinV-FITC(Fig. 9 )
Evaluation of nanocomplex-induced cytotoxicity in B16F10 cells upon exposure to 405 nm or 532 nm wavelengths as assessed by MTT and cell cycle analysis
Viability of B16F10 cells exposed to MNPs-CHI-TPP-TRAIL, MNPS-CHI-TPP-CUR, or both. Then exposed to magnetofection for 15 min and light (wavelengths 405 nm or 532 nm) after 24 h. Cell viability was measured by MTT assay (A). Effect of MNPs-CHI-TPP-TRAIL, MNPS-CHI-TPP-CUR on cell cycle in B16F10 cells. 1 × 106 cells were seeded in a 6-well plate, and cells were exposed to light (wavelengths 405 nm or 532 nm) after 24 h. Cell-cycle analysis was measured by flow cytometry (B). Data were expressed as mean ± SD. Statistical analyses were performed by second-way ANOVA followed by Tukey. Symbols indicate significance: p < 0.01 (*), p < 0.001 (**), and p < 0.0001 (***) differences between no light, 405 nm, and 532 nm treated samples
Discussion
In this study, we obtained nanoparticles with a diameter of ~ 206.2 nm and a polydispersity index (PdI) of 0.192 (Fig. 3A). Particle size can affect the biodistribution, efficiency (number of nanocomplexes found within cells at a given time), and pathway of cell uptake by influencing their adhesion and interaction with cells [21]. On the other hand, the PdI is basically a representation of the size distribution of a given sample, whose values vary from 0.0 (shows perfectly uniform with respect to the particle size) to 1.0 (highly polydispersity sample with populations including particle of multiple sizes) [22]. PdI values of 0.2 and lower are considered optimal for nanoparticles according to their polymeric materials [23], and on this basis, a homogeneous sample of nanocomplexes consisting of chitosan was obtained in this work as indicated by pDNA release assay. Chitosan-based magnetic pDNA nanocomplexes had a positive zeta potential of 19.6 mV in 100% of the sample (Fig. 3B). According to the literature, because of the negative charge characteristic of the cell membrane, cationic nanocomplexes are internalized with greater efficiency than nanocomplexes with anionic or neutral surface loads [24]. Regarding the DNA retention profile, 80% retention and progressive release until complete release after 24 h was observed (Fig. 4). Controlled drug release systems are commonly used to optimize the therapeutic effect of medical treatment, reduce side effects, maintain the drug at an optimal concentration for long periods of time and decrease the frequency of drug administration [25]. Regarding the cytotoxic effect of curcumin, an LD50 of 4.0719 µM (~ 1.5 µg/mL) at 24 and 48 h and an LD50 of 1.018 µM (~ 0.375 µg/mL) at 72 h were obtained, and the mean lethal dose of curcumin (Fig. 5) indicated that it will have a time-dependent effect on tumor cells.
Nanocomplexes with curcumin based on chitosan were observed by electronic transmission microscopy (TEM) (Fig. 6A). The vast majority of the nanocomplexes used for drug shipments are spherical in shape; however, other forms, such as cubes, cylinders, ellipsoids, and discs, have been proposed as novel drug carriers [14].The results of the Zetasizer reader, the nanocomplexes were 258.6 nm in diameter with a PdI of 0.110 (Fig. 6B), which are optimal values because they are in the range of optimal in vitro phagocytosis and optimal homogeneity, respectively [23, 26]. The difference in size observed by the Zetasizer readings and TEM analysis was due to the characteristics of the sample at the time of the reading. On the other hand, a Zetasizer requires the sample of nanocomplexes dispersed in solution, while for TEM, a dry flat surface sample is used, and the difference in the sample can influence the morphology of the nanocomplex due to the flexible nature of the chitosan polymer chitosan-based magnetic curcumin nanocomplexes showed a positive zeta potential of 30.3 mV in 100% of the sample.
The observed retention of the nanocomplexes (Fig. 7) was approximately 55%, while other studies showed encapsulation efficiencies from 61.22 to 73.44% [27]. The low encapsulation efficiency may have been due to the hydrophobicity of the curcumin and the molecular weight of the chitosan used. The literature indicates that long chains of high-molecular-weight chitosan can trap larger amounts of curcumin during ion gelation with TPP [19,20,21,22,23,24,25,26,27,28]; however, increasing the concentration of curcumin in the formulation decreased the encapsulation efficiency of the final nanocomplexes. The release after standardization is progressive, and the highest release was observed at 12 h, while at 24 h, an almost complete release was observed. Similar to the chitosan-based nanocomplexes with DNA, the progressive release of curcumin allows the optimization of the therapeutic effect, among the other aforementioned effects [25].
To the analysis of the morphology of the B16F10 cells were exposed to nanocomplexes. The corresponding control cells were not treated. Treatment with vehicle (MNPs-CHI-TPP) showed a little noticeable effect on cell morphology. In the case of the sample treated with MNPs-CHI-TPP-TRAIL, the cells presented with different coloration; however, the elongated morphology was maintained. On the other hand, treatment with MNPs-CHI-TPP-CUR for 24 h led to an obvious morphology change in the cells, which presented with a circular morphology. The image of the cells treated with MNPs-CHI-TPP-CUR + MNPs-CHI-TPP-TRAIL for 24 h appeared thinner and darker compared to the control cells. After 48 and 72 h of treatment, noticeable morphological changes and decreased confluence was observed (Fig. 8A). The efficiency of the transfection using magnetic nanoparticles with chitosan, as well as the expression of TRAIL-induced by the application of an external magnetic field, has been demonstrated in other works carried out in the laboratory [29, 30].
Also, has been demonstrated that TRAIL (Apo2L), is a protein of the family TNF (tumor necrosis factor), highly selective for the elimination of tumor cells by induction of apoptosis, with slight toxicity to somatic cells [14]. TRAIL interacts with four membrane-anchored receivers: DR4, DR5, DcR1, and DcR2. DR4 and DR5 receptors contain a demonized conserved cytoplasmic region “death domain,” which is required for TRAIL-mediated apoptosis induction. On the other hand, DcR1 lacks an intracellular domain while DcR2 contains a truncated death domain, so both receptors (DcR1 and DcR2) could serve as decoys and protect the induction cells from TRAIL-mediated apoptosis [18].
Cells treated with the MNPs-CHI-TPP-CUR nanocomplexes showed decreased viability with respect to the control cells. After 24 h of treatment, viability of 91.61% was observed with respect to the control; after 48 h, this viability decreased to 86.43%, and finally, after 72 h, it decreased to 72.22% (Fig. 8B). The statistical analysis showed no significant difference between cells treated with MNPs-CHI-TPP-CUR nanocomplexes and the control. Cultured cells treated with both types of nanocomplexes (MNPs-CHI-TPP-TRAIL and MNPs-CHI-TPP-CUR) showed a 24-h decrease in viability of 30% with respect to the control. After 48 h of treatment, there was a decrease in viability of approximately 70% with respect to the control and a significant difference with respect to all treatments. Finally, after 72 h, there was a decrease in cell viability of more than 40% with respect to the control. This result indicates a clear potentiating effect of curcumin in the induction of TRAIL-mediated apoptosis. Other studies have been evaluated the cytotoxic effect of chitosan-based nanocomplexes with curcumin and TRAIL expression vectors without magnetism induction in HCT116 cells, and their results showed a greater decrease in cell viability with a synergistic effect of TRAIL and curcumin than was observed for each nanocomplex type administered individually [11].
The morphology of the B16F10 cells exposed to each treatment and specific wavelengths of 405 nm and 532 nm were analyzed. The B16F10 cells of the control group exposed at a wavelength of 405 nm (100 mW) had an elongated morphology and high confluence in addition to a lack of internal vesicles and granules, indicating a null effect of exposure for 15 min. Cells treated with MNPs-CHI-TPP-TRAIL and wavelength at 405 nm (100 mW) exhibited decreased confluency and cell agglomerations. Cells treated with MNPs-CHI-TPP-CUR with and wavelength of 405 nm (100 mW) showed an obvious decrease in confluence in addition to a circular morphology with the elongated morphology was completely lost. Finally, cells treated with both MNPs-CHI-TPP-TRAIL and MNPs-CHI-TPP-CUR nanocomplexes at a wavelength of 405 nm (100 mW) showed loss of confluence and loss of cell morphology. Cells exposed to a wavelength of 532 nm (100 mW) for 15 min. in the control group were highly confluent and were elongated with semi-epithelial morphology, which indicated a small or negligible irradiation effect. Treatment with MNPs-CHI-TPP-TRAIL led to slight changes in confluence and no drastic morphological change. On the other hand, cells treated with MNPs-CHI-TPP-CUR exhibited a decrease in cell confluence, and although cells with circular morphology were not observed, cell clusters with irregular morphologies were observed. Cells treated with both types of nanocomplexes exposed at 532 nm (100 mW) showed lower confluence, compared to the control, as well as irregular clusters. Death cellular decreased markedly 24 h after each of the irradiated treatments, regardless of the wavelength used (405 nm or 532 nm for 15 min) with respect to the control. Previously data indicated that the cytotoxic effect of curcumin in tumor cells can be enhanced by visible light or blue light at wavelengths close to the maximum absorbance of curcumin [31]. Finally, cells exposed to both nanocomplexes and exposed to wavelengths of 405 nm or 532 nm showed significant differences compared to those exposed to nanocomplexes without light. Therefore, the effect of the MNPs-CHI-TPP-TRAIL and MNPs-CHI-TPP-CUR nanocomplexes exposed to wavelengths of 405 nm and 532 nm was synergistic and enhanced by the generation of reactive oxygen species induced by curcumin that produce an increased endosome release molecules; there is a possible explanation for how the curcumin photoactivation may enhance the TRAIL gene function due to the ability to increase transfection when curcumin is exposed to laser light [32] (Fig. 10A). In the cell cycle analysis (Fig. 10B), Subg0/1 indicates DNA degradation, and the arrest in the G1 stage indicates that cells are preparing for apoptosis [33]. For that reason, we can conclude that our treatment is working to induce DNA degradation and possible apoptosis of the B16F10 cell line.
The explanation of the synergistic effect between TRAIL and CUR in nanoparticles for apoptosis induction is shown in Fig. 11.
Conclusions
In this work, MNPs-CHI-TRAIL and MNPs-CHI-CUR were synthesized, and magnetic core was made by chemical reduction and coated with chitosan by ionic gelation modifications. The analysis showed that the nanocomplexes were monodisperse and stable. The combination of nanocomplexes containing a TRAIL gene and curcumin drug exhibited a synergistic effect on cell death induction of murine cancer cells. The effect of the combination of nanoparticles was enhanced by exposing the transfected culture to laser photoactivation at wavelengths 405 and 532 nm. This is the first approach for synergic interaction between a TRAIL gene and curcumin drug to improve the apoptosis in cancer cells. These data support the possibility to developing apoptosis induction strategies in cancer-based on the combination of the TRAIL gene and the curcumin molecule and using the photoactivation property of this molecule to develop new chemogenephotodynamic therapies.
Abbreviations
- MNPs:
-
Magnetic nanoparticles
- CUR:
-
Curcumin
- TPP:
-
Sodium tripolyphosphate
- CHI:
-
Chitosan
References
Hiremath CG, Heggnnavar GB, Kariduraganavar MY et al (2019) Codelivery of paclitaxel and curcumin to foliate positive cancer cells using Pluronic-coated iron oxide nanoparticles. Prog Biomater 8:155–168
Mirnejad R, Mofazzal Jahromi M, Al-Musawi S, Pirestani M, Fasihi Ramandi M, Ahmadi K, Rajayi H, Mohammad Hassan Z, Kamali M (2014) Curcumin-loaded chitosan tripolyphosphate nanoparticles as a safe, natural and effective antibiotic inhibits the infection of Staphylococcus aureus and Pseudomonas aeruginosa in vivo. Iran J Biotechnol 12(3):1–8
Khandelwal P, Alam A, Choksi A, Chattopadhyay S, Poddar P (2018) Retention of anticancer activity of curcumin after conjugation with fluorescent gold quantum clusters: an in vitro and in vivo xenograft study. ACS Omega 3(5):4776–4785
Goel A, Kunnumakkara AB, Aggarwal BB (2008) Curcumin as “curcumin”: from kitchen to clinic. Biochem Pharmacol 75(4):787–809
Justin C, Samrot AV, P DS, Sahithya CS, Bhavya KS and Saipriya C, (2018) Preparation, characterization, and utilization of core shell super paramagnetic iron oxide nanoparticles for curcumin delivery. PLoS ONE 13(7):e0200440. https://doi.org/10.1371/journal.pone.0200440
Roos F, Binder K, Rutz J, Maxeiner S, Bernd A, Kippenberger S, Zöller N, Chun FK, Juengel E, Blaheta RA (2019) The antitumor effect of curcumin in urothelial cancer cells is enhanced by light exposure in vitro. Evid Based Complement Alternat Med 11:6374940. https://doi.org/10.1155/2019/6374940
de Matos RPA, Calmon MF, Amantino CF, Villa LL, Primo FL, Tedesco AC, Rahal P (2018) Effect of curcumin-nanoemulsion associated with photodynamic therapy in cervical carcinoma cell lines. BioMed Res Int :4057959. https://doi.org/10.1155/2018/4057959.
Trigo Gutierrez JK, Zanatta GC, Ortega ALM et al (2017) Encapsulation of curcumin in polymeric nanoparticles for antimicrobial Photodinamic Therapy. PLoS ONE 12(11):e0187418. https://doi.org/10.1371/journal.pone.0187418
Kamel AE, Fadel M, Louis D (2019) Curcumin-loaded nanostructured lipid carriers prepared using Peceol™ and olive oil in photodynamic therapy: development and application in breast cancer cell line. Int J Nanomed 14:5073–5085
Yang X, Li Z, Wu Q, Chen S, Yi C, Gong C (2017) TRAIL and curcumin codelivery nanoparticles enhance TRAIL-induced apoptosis through upregulation of death receptors. Drug Delivery 24(1):1526–1536
Shankar S, Chen Q, Sarva K, Siddiqui I, Srivastava RK (2007) Curcumin enhances the apoptosis-inducing potential of TRAIL in prostate cancer cells: molecular mechanisms of apoptosis, migration, and angiogenesis. J Mol Signal 2(10).https://doi.org/10.1186/1750-2187-2-10
Duse L, Agel MR, Pinnapireddy SR, Schäfer J, Selo MA, Ehrhardt C, Bakowsky U (2019) Photodynamic therapy of ovarian carcinoma cells with curcumin-loaded biodegradable polymeric nanoparticles. Pharmaceutics 11(6):282. https://doi.org/10.3390/pharmaceutics11060282
Park S, Cho DH, Andera L, Suh N, Kim I (2013) Curcumin enhances TRAIL-induced apoptosis of breast cancer cells by regulating apoptosis-related proteins. Mol Cell Biochem 383(1–2):39–48
Alvizo-Baez CA, Luna-Cruz IE, Vilches-Cisneros N, Rodríguez-Padilla C, Alcocer-González JM (2016) Systemic delivery and activation of the TRAIL gene in lungs, with magnetic nanoparticles of chitosan controlled by an external magnetic field. Int J Nanomed 11:6449–6458
Jung EM, Lim JH, Lee TJ, Park JW, Choi KS, Kwon TK (2005) Curcumin sensitizes tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)- induced apoptosis through reactive oxygen species-mediated upregulation of death receptor 5 (DR5). Carcinogenesis 26(11):1905–1913
Yang Xi, Li Z, Qinjie Wu, Chen S, Yi C, Gong C (2017) TRAIL and curcumin codelivery nanoparticles enhance TRAIL-induced apoptosis through upregulation of death receptors. Drug Delivery 24(1):1526–1536
Lehmann J, Agel MR, Engelhardt KH, Pinnapireddy SR, Agel S, Duse L, Preis E, Wojcik M, Bakowsky U (2021) Improvement of pulmonary photodynamic therapy: nebulisation of curcumin-loaded tetraether liposomes. Pharmaceutics 13(8):1243
Rodríguez-de la-Fuente AO, Alcocer-González JM et al (2009) Effect of 60 Hz electromagnetic fields on the activity of Hsp70 promoter: an-in vitro study. Cell Biol Int 33:419–433
Calvo P, Remuñan-López C, Vila-Jato JL, Alonso MJ (1997) Novel hydrophilic chitosan-polyethylene oxide nanoparticles as protein carriers. J of App Pol Sci 63(1):125–132
Yang H, Masse S, Zhang H, Hélary C, Li L, Coradin T (2014) Surface reactivity of hydroxyapatite nanocoatings deposited on iron oxide magnetic spheres toward toxic metals. J Colloid Interface Sci 417:1–8
Shapero K, Fenaroli F, Lynch I, Cottell DC, Salvati A, Dawson KA (2011) Time and space resolved uptake study of silica nanoparticles by human cells. Mol BioSyst 7(2):371–378
Danaei M, Dehghankhold M, Ataei S, Hasanzadeh Davarani F, Javanmard R, Dokhani A, Mozafari M (2018) Impact of particle size and polydispersity index on the clinical applications of lipidic nanocarrier systems. Pharm 10(2):57. https://doi.org/10.3390/pharmaceutics10020057
Salatin S, Yari KA (2017) Overviews on the cellular uptake mechanism of polysaccharide colloidal nanoparticles. J Cell Mol Med 21(9):1668–1686. https://doi.org/10.1111/jcmm.13110
Banquy X, Suarez F, Argaw A, Rabanel JM, Grutter P, Bouchard JF, Giasson S (2009) Effect of mechanical properties of hydrogel nanoparticles on macrophage cell uptake. Soft Matter 5(20):3984–3991
Heng PWS (2018) Controlled release drug delivery systems. Pharm Dev Technol 23(9):833–833. https://doi.org/10.1080/10837450.2018.1534376
Biswas A, Islam MR, Choudhury ZS, Mostafa A and Kadir MF (2014) Nanotechnology based approaches in cancer therapeutics. Adv Nat Sci: Nanosci Nanotechnol 5(4):04300. https://doi.org/10.1088/2043-6262/5/4/043001.
Kumara JB, Madhusudhan B (2015) Synthesis, characterization and hemocompatibility evaluation of curcumin encapsulated chitosan nanoparticles for oral delivery. Int J Adv Res 3(4):604–611
Wu Y, Yang W, Wang C, Hu J, Fu S (2005) Chitosan nanoparticles as a novel delivery system for ammonium glycyrrhizinate. Int J Pharm 295(1–2):235–245
Alvizo Báez CA, Luna Cruz IE, Rodríguez Padilla C, Alcocer González JM (2014) Magnetic nanoparticles of chitosan for targeted delivery system of plasmids to the lungs. J of Nanotechnology 14. https://doi.org/10.1155/2014/313415
Degli-Esposti MA, Dougall WC, Smolak PJ, Waugh JY, Smith CA, Goodwin RG (1997) The novel receptor TRAIL-R4 induces NF-κB and protects against TRAIL-mediated apoptosis yet retains an incomplete death domain. Immunity 7(6):813–820
Bernd A (2014) Visible light and/or UVA offer a strong amplification of the antitumor effect of curcumin. Phytochem Rev 13(1):183–189
Pinnapireddy SR, Duse L, Akbari D, Bakowsky U (2017) Photo-enhanced delivery of genetic material using curcumin loaded composite nanocarriers. Clin. Oncol, 2: 1323. https://doi.org/10.25107/2474-1663.1323.
Lin QH, Que FC, Gu CP, Zhong S, Zhou D, Kong Y, Yu L, Liu SW (2017) ABT-263 induces G1/G0-phase arrest, apoptosis and autophagy in human esophageal cancer cells in vitro. Acta Pharmacol Sin 38(12):1632–1641
Funding
This study is funded by Grant PAICYT CN 1179–20 Universidad Autónoma de Nuevo León.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study’s conception and design. Material preparation, data collection, and analysis were performed by Cynthia Aracely Alvizo Báez, Arnulfo Aramis Peña-Torres, Luis Daniel Terrazas-Armendáriz, Itza Eloisa Luna-Cruz, Ashanti Concepción Uscanga Palomeque, Adriana Sampayo-Reyes, Reyes S. Tamez-Guerra, Cristina Rodríguez-Padilla, and Juan Manuel Alcocer-González. The first draft manuscript was written by Cynthia A Alvizo Báez, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interest.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Alvizo-Báez, C.A., Peña-Torres, A.A., Terrazas-Armendáriz, L.D. et al. Synergic effect between TRAIL gene and curcumin in magnetic chitosan nanoparticles on cancer cells apoptosis enhanced by laser photoactivation. J Nanopart Res 24, 165 (2022). https://doi.org/10.1007/s11051-022-05544-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11051-022-05544-x