Abstract
Candida auris is responsible for hospital outbreaks worldwide. Some C. auris isolates may show concomitant resistance to azoles, echinocandins, and polyenes, thereby possibly leaving clinicians with few therapeutic options. In addition, this multi-drug-resistant yeast is difficult to identify with conventional methods and has the ability to persist on environmental surfaces causing hospital-acquired infections. The development of new treatment options and tools for identification is critical to control, prevent, and establish an early diagnosis of this emerging pathogen. The aim of this study was to perform a critical patent review to explore and identify the latest advances in therapeutic strategies as well as diagnostic methods for C. auris. A total of 19 patents were identified for a preliminary assessment from the Espacenet database. Three patents were excluded as they were out of focus for this review according to their abstract and/or description. The final selection covered 16 patents, which were surveyed by country, year and classified as treatment or diagnostic methods for C. auris. As noted in the patent reading, in recent years, the interest of academic, government and industry sectors have shown an increasing tendency focused on research and development of new therapeutic molecules and diagnostic methods to combat this emerging pathogen.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Candida auris is an emerging fungus that presents a serious global health threat. Though C. auris was first identified in Japan in 2009 [1], a retrospective study revealed that the earliest strain of C. auris dates back to 1996 in South Korea [2], and since then, outbreaks have been reported from over 47 countries [3].
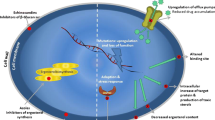
Controlling C. auris is a major concern for several reasons: (1) It is resistant to multiple classes of antifungal drugs commonly used to treat Candida infections, (2) It is difficult to identify with standard laboratory methods and thus can often be misidentified without the use of specific technology leading to inappropriate management and (3) It can disseminate between patients in healthcare settings. For these reasons, rapid identification of clinical C. auris strains is very important so that healthcare facilities can take necessary measures to prevent its transmission [2, 4]. In addition, C. auris is exceptionally well adapted to the nosocomial environment, resists common disinfectants, persists on medical equipment and dry surfaces in hospitals for up to 4 weeks, and readily colonizes the axilla, groin, and nares of patients [5].
Ultimately, correct detection and identification of the pathogen along with its antifungal susceptibility profile, followed by strict adherence to appropriate treatment and infection prevention and control strategies, are crucial for limiting the spread of C. auris [6]. Furthermore, matrix-assisted laser desorption ionization-time of flight mass spectrometry ("MALDI-TOF MS) and ribosomal DNA sequencing are preferred over conventional diagnostic methods as they can reliably distinguish C. auris from other yeasts [6].
Currently, four classes of antifungal agents are available for the treatment of candidiasis: azoles (fluconazole, voriconazole), polyenes, echinocandins (caspofungin, micafungin, and anidulafungin), and flucytosine. The use of echinocandins as first-line therapies subjected to sensitivity testing is recommended [7]. Although echinocandins are effective against most isolates of Candida species resistant to other antifungal agents, an increase in the number of C. auris strains with reduced susceptibility to one or more echinocandins has been observed [8, 9].
All these factors allow C. auris to easily spread in hospitals and cause recalcitrant outbreaks. They also contribute to the high mortality rates (30–60%) observed in the case of C. auris invasive infections [10]. The onset of the SARS-CoV-2 pandemic has increased the C. auris colonization and candidemia cases [4]. New C. auris outbreaks in critically ill COVID-19 patients were also reported, and it was alerted that the SARS-CoV-2 pandemic might facilitate the transmission of nosocomial pathogens, including C. auris[11].
Thus, this review aims to provide an overview of new methods and therapeutic options that can bring new hope to patients and healthcare professionals to combat the outbreaks caused by C. auris.
Methods
An updated keyword search of “Candida” and “auris” was performed in the claim search field of Espacenet database on May 30, 2022. This review is based on the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines. The patent selection was based on the following inclusion criteria: recent patents published after the first description of C. auris in 2009 in Japan to May 2022 in any language and containing “Candida” and “auris” as the keywords in the title or abstract. Using the same criteria, a literature survey was performed using the PubMed literature database to compare the number of papers on the subject of “Candida auris” to the number of patents found in the patent databases. This survey was conducted in May 2022. Patents that did not focus on the “Candida” and “auris” subject were excluded from the search results. A total of 21 patents were selected for preliminary assessment from Espacenet (Fig. 1). After excluding three patents that were out of scope, 18 patents were selected to be studied in detail. Following this, the patent contents were classified per country, year and type of applicant.
Results and Discussion
The identification of new target-specific antifungal therapeutic strategies and the development of novel diagnostic methods against C. auris are valuable approaches to combat this emerging multidrug-resistant pathogen. In our review, the patents related to the diagnosis and treatment published by May 2022 were included. Out of 18 reviewed patents, 11 were deposited in 2020, and until May 2022, 4 were described. C. auris was first reported in 2009 in Japan, but patents related to C. auris appeared nine years later. The total number of patent publications between 2018 and 2020 was also lower compared to the number of scientific articles published during the same period. Using the same combination of words "Candida auris” to search in the PubMed database, 882 scientific papers were obtained from 2009 to 2022 (accessed on May 23, 2022). Based on the results, we observed that both the number of patents and the number of scientific publications increased rapidly, emphasizing the importance of C. auris as a global health concern. We have to considerer the impact of the COVID-19 pandemic on the patent community. Most of the offices suspended the transmittal of Patent Cooperation Treaty (PCT) documents, limited, or canceled in-person interviews, postponed deadlines, and focused only on patents related to the COVID-19 pandemic [12]. Quarantines and lockdowns have shut down conferences, seminars, and other social events where knowledge is disseminated, collaboration occurs, funding sources are identified, and discoveries emerge. In addition, many clinical trials and early-stage research projects that could lead to life-saving treatments worldwide have been abandoned due to lockdowns or because funding has been discontinued [13]. National Public Radio (NPR) reported an interruption of clinical trials for many important cancer drugs [14]. At present, only 4 clinical studies are registered in the Clinical Trials related to C. auris (accessed on May 23, 2022: https://clinicaltrials.gov). In the USA, non-COVID-19 research operations at universities, medical schools, and federal labs (most located in states with severe lockdowns) have also been shut down, leading to the cancellation of basic and applied research on several diseases that kill millions each year [15].
Regardless of the countries which produced patents, the US was responsible for the majority of the number of patents published (n = 10), followed by China (n = 6), Spain (n = 1) and Japan (n = 1) (Fig. 2). Though C. auris is a global concern causing outbreaks in more than 48 countries (Fig. 2), the patents are mostly based on cases of two countries. Although the US has presented a high number of patent publications, owing to its excellent economy and investment in the field of technological innovation [16] in the past two decades, China has experienced strong and sustained growth in patent applications, surpassing the US and becoming the world’s leading patent registrar [17].
Several scientific organizations may request the protection of an invention by filing a patent application. Industries lead the filing ranking with 11 patents (61.1%). In addition, hospitals (02/18, 11.1%), universities (02/16, 11.1%), universities in collaboration with research centers (01/18, 5.5%), only research centers (01/18, 5.5%) and independent researcher (01/18, 5.5%) also file for patents applications.
Based on our findings, the main focus of the patents was categorized into treatment (Table 1) and diagnostic methods (Table 2). Table 1 represents patent publications that focus on the C. auris treatment/disinfection. Table 2 describes new methods to identify and diagnose C. auris colonization and infection. The treatment and diagnostic methods are discussed in the following sections in detail:
Treatment of C. auris Infection or/Colonization
In addition to antifungal resistance, C. auris has a special predilection for skin, particularly the axilla and groin. It has the potential to colonize hosts within days to weeks of exposure, and invasive infections may occur within days to months after colonization [10]. Colonization with C. auris may persist for many months and possibly indefinitely [4]. This information is helpful for better treatment of patients.
“WO2020232037 (A1)” [18] describes the use of Ibrexafungerp (formerly known as SCY-078) as the first compound of the enfumafungin-derived triterpenoid class of (1 → 3)-β-D-glucan synthase inhibitors (GSIs). There is also an open-label study registered in the clinical trials (https://clinicaltrials.gov/ct2/home) to evaluate the efficacy, safety, tolerability, and pharmacokinetics of oral Ibrexafungerp (SCY-078) as an emergency treatment of patients with candidiasis and candidemia caused by C. auris. After oral administration, the enfumafungin derivatives were able to significantly reduce C. auris infection of the skin, revealed a useful strategy to prevent transmission, and limited the risk of C. auris outbreaks [36, 37]. Increased bioavailability of Ibrexafungerp positions them to be an optimal solution for the decolonization of C. auris from anatomic areas of a subject [38]. On the other hand, glucan synthase inhibitors, such as echinocandins, have a similar mechanism of action and lower oral bioavailability, and their target site concentrations are much lesser than that of plasma [39]. Thus, they cannot prevent the colonization of C. auris at multiple body sites, including nares, groin, axilla, and rectum for 3 months or more after initial intravenous echinocandin treatment [3]. In addition, Ibrexafungerp does not have clinically relevant antibacterial properties and cannot cause a deleterious effect on the normal bacterial microbiome of the skin and mucosa. Ibrexafungerp, with the potential to provide the therapeutic advantages of both intravenous (IV) and oral formulations, being developed as the first oral and intravenous glucan synthase inhibitor (IV GSI) for the treatment and prevention of fungal infections, including serious and life-threatening infections due to Candida spp., Aspergillus spp. and Pneumocystis jirovecii [40]. Ibrexafungerp causes a decrease in the concentration of (1 → 3)-β-D-glucan polymers and weakens the fungal cell wall [37].
“WO2021090739 (A1)” [19] describes a pharmaceutical composition comprising of a therapeutically effective amount of 4-[16]propyl)-1-piperidinyl]propoxy}benzamidine (abbreviated as T-2307) which is effective in treating C. auris infection. Results demonstrate that T-2307 is effective against invasive infections caused by Candida spp., specifically C. auris, as both in vitro and in vivo activity were observed against this emerging pathogen. The initial preclinical and early-stage clinical development of T-2307 has been conducted by FUJIFILM Toyama Chemical Co. Ltd. (Tokyo, Japan), which has recently assigned the rights to Appili Therapeutics (Halifax, NS, Canada; ATI-2307) (http://fftc.fujifilm.co.jp/en/news/news191121e.html, accessed on July 07, 2021) for developing and marketing the drug outside of Japan. In vitro study using clinical isolates of C. auris showed that the MICs of T-2307 ranged from ≤ 0.008 µg/mL to 0.015 µg/mL, and the drug was active against strains that were resistant to fluconazole [41]. This agent has a novel mechanism of action and causes the collapse of the mitochondrial membrane potential. Recent studies have revealed that this activity is selective for fungi [41].
“WO2020150532 (A1)”[20] refers to a composition comprising of polyhexanide biguanide (PHMB) for the topical treatment of mucous membranes or skin infected with C. auris. PHMB alone or in combination with one or more cationic biocides as polyaminopropyl biguanide (PAPB) and/or chlorhexidine (CHG) presented activity against C. auris. The results showed that the treatment with the formulation resulted in a significant decrease in the colonization of C. auris on murine ear surfaces. The mechanism of action consists of disrupting microbial cell membranes and metabolism, interfering with function, and ultimately destroying the microbial cell [42].
“US2020237705 (A1)” [21] refers to glycerol monolaurate (GML), also known as monolaurin, which kills C. auris. In addition, 5% GML gel was able to inhibit biofilm formation of C. auris. There is no published study at present confirming the antifungal activity of GML against C. auris. However, oral topical treatments of GML have resulted in a significant decrease in colony formation unit of C. albicans on tongue tissue compared to the vehicle control. The results show that GML is a promising antifungal compound in vivo and can be used in the future for the treatment of oral candidiasis [43]. There is no mechanism of action described for GML.
“CN111954533 (A)”[22] comprising of taurolidine, refers to a method for treating C. auris in blood. When the drug reaches the infection site of C. auris from the point of entry, the hydrolysable polymer coating covering the taurolidine core acts as a sacrificial layer and slowly decomposes as the nanoparticles pass through the bloodstream. Eventually, the hydrolysable polymer coating decomposes to the point where the taurolidine core is exposed to blood. The taurolidine nucleus is then hydrolyzed into its active part (hydroxymethyl derivative), which is then targeted to C. auris infection. In addition, clinical trials with taurolidine have been conducted to prevent infections in catheters [44]. The mechanism of taurolidine anti-adherence activity is not known.
“WO2018204506 (A1)”[23] refers to cationic steroidal antimicrobials (“CSAs”) and their formulations used for the treatment of fungal infections and colonization on medical devices. CSA-131 showed activity against all four clades of C. auris with no variation in activity between the clades. The MIC value distribution of CSA-131 ranged from 0.5 to 1 mg/L [45]. Membrane disruption has been identified as a major mechanism of antifungal activity of CSAs [45].
“US2021030852 (A1)” [24] refers to an immunizing method against C. auris infection using agglutinin-like sequence-3 (Als3). It was discovered that C. auris harbors homologs of C. albicans Als cell surface proteins. It was found that C. albicans NDV-3A vaccine, harboring the N-terminus of Als3p formulated with alum, generates cross-reactive antibodies against C. auris clinical isolates and protects neutropenic mice from hematogenous disseminated C. auris infection. In addition, the NDV-3A vaccine also displayed a protective effect in neutropenic mice when combined with micafungin [46]. A recent study demonstrated that the vaccine was predicted to be stable, soluble, antigenic, non-allergic with desirable physicochemical properties. The results showed the candidate vaccine as a promising alternative therapy for the treatment of C. auris [46].
“WO2021178774A1” [25] refers to a combination between hydrogen peroxide plus acetic acid to form peroxy acetic acid to disinfect surfaces contaminated by C. auris. Appropriate cleaning practices are also key to the management of C. auris infection, as commonly utilized disinfectants are not guaranteed to be active against C. auris. The Environmental Protection Agency (EPA) has a list of registered, hospital-grade disinfectants with activity against C. auris, including hydrogen peroxide/peroxyacetic acid (Selected EPA-Registered Disinfectants. Available online: https://www.epa.gov/pesticide-registration/selected-epa-registered-disinfectants#candida-auris (accessed on 30 May 2022). However, higher concentrations of this disinfectant coupled to longer exposure times were required to lower regrowth, but even then, they were not able to completely eradicate the pathogen [47]
New Diagnostic Methods
Misidentification of C. auris with other yeasts (e.g., C. haemulonii, C. famata, C. guilliermondii, C. lusitaniae, C. parapsilosis) may occur due to the use of standard biochemical methods and commercially available tests [6]. In fact, its correct identification at the species level requires more advanced techniques, such as DNA sequencing MALDI-TOF MS or both. Whether the microorganism has been isolated from sterile and non-sterile body sites, it is observed that asymptomatic colonization represents a risk for C. auris transmission [48].
“WO2020114998 (A1)” [26] provides primers and probes to identify C. auris. It includes oligonucleotide primers and fluorescent-labeled hydrolysis probes that hybridize to a specific target within the C. auris genome. This region can be specifically identified using TaqMan® amplification and detection method. As a result of the analysis, a C. auris target was chosen, which was 5.8S/ITS2 rRNA gene (GenBank accession number AB375772). The disclosed methods may include performing at least one cycling step using one or more pairs of primers that includes amplifying one or more portions of the target gene of the nucleic acid molecule from a sample.
“CN110951905 (A)” [27] and “CN112041441 (A)”[33] provides a method for quick and accurate detection of C. auris. Both comprises a primer set and a detection kit used to amplify and detect the target sequence of C. auris in the test body by the by the loop-mediated isothermal amplification (LAMP) method. LAMP approach was proven to reliably identify all of the tested C. auris strains, distinguishing these isolates from other species (even very closely related species) with a 100% specificity. The results were obtained within a short time, without any technical complications related to the use of the amplification instrument [49]. This technique is expected to save the time of the clinicians required for cultivation and DNA extraction, thus allowing an early diagnosis. Portable LAMP amplification equipment has been made commercially available. Overall, this assay should be particularly valuable for C. auris, a pathogen that is an important target of environmental control in health care facilities[49].
“CN110408720 (A)” [28] was developed in order to provide a method for the identification of C. auris. With the help of a high-resolution melting curve C. auris can be identified and distinguished from other similar species. Signature melting profiles were generated for C. auris, C. duobushaemulonii, C. haemulonii, and C. lusitaniae, enabling their unambiguous discrimination. Excellent results were achieved with assays during the development phase, as well as during the proficiency panel validation [50].
“ES2763043 (A1); ES2763043 (B2)”[29] describes a biosensor for the detection of C. auris DNA and/or diagnosis of infection caused by C. auris. The biosensor is based on a porous material comprising of a reporter molecule and single-stranded DNA oligonucleotides that specifically recognize a particular region of C. auris DNA. Thus, when the DNA of C. auris is present in the medium, the oligonucleotides recognize the said region, bind to it, and the reporter is released, which is then detected. Recently, the detection of C. auris by using a fluorogenic nanosensor has been reported [51]. A nanoporous anodic alumina scaffold is filled with fluorescent indicator rhodamine B and the pores are blocked with different oligonucleotides capable of specifically recognizing C. auris genomic DNA. C. auris is detected even at low concentrations, thus allowing to obtain a diagnostic result in clinical samples within an hour with no prior DNA extraction or amplification steps [51].
“US2020291488 (A1)” [30] provides a method to determine if C. auris is present in a biological or environmental sample. An assay for a rapid detection method based on the culture-independent T2 Magnetic Resonance (T2MR) technology has been used to detect C. auris from whole blood and common swab matrices at concentrations < 10 CFU/mL. Testing with clinical samples indicates that this test can be used to identify C. auris and other species from patient blood samples without the requirement of blood culture. This rapid and sensitive test enables the detection of C. auris in candidemia patients and assists in the screening, isolating, and monitoring of the spread of this emerging multidrug-resistant pathogen. In addition, the T2MR method can be used to detect C. auris from clinical skin swab samples [52].
“US10870829 (B2); US2020270567 (A1)” [31] refers to a system to identify C. auris which is based on two aspects related to the sensitivity to a different medium. The first is the positive selection of C. auris based on its distinctive resistance to quaternary ammonium compounds (especially at elevated incubation temperatures). The second is the negative selection of C. auris based on its distinctive sensitivity to tert-Butyl-hydroperoxide. C. auris can be identified in a sample by the use of a positive-selection culture medium which fosters C. auris colony growth while suppressing the growth of other yeasts. The isolate can be confirmed as C. auris by using a negative-selection culture medium that suppresses C. auris colony growth while permitting the growth of other yeasts. There is no published study describing this method to date.
“CN110551840 (A)” [32] refers to a nucleic acid reagent, kit, system, and method for the detection of invasive fungi. The nucleic acid reagent comprises specific primers and a specific code probe sequence. At least nine invasive fungi, including C. auris, can be detected, resulting in a fast, comprehensive, sensitive, specific, and automatic detection.
“CN110804671 (A)”[34] proposes the use of rDNA genes as biomarkers for C. auris, which includes its ITS1-ITS4 nucleotide sequence. It was found that the rDNA genes ITS1-ITS4 of C. auris are conserved, and some of the sequences are specific and can be used as ideal primers and templates for the study of nucleic acid-based detection of C. auris. Using the rDNA gene (SEQ IDNO:17) sequence as a biological target to establish a real-time fluorescent quantitative PCR detection method for C. auris will help in the rapid diagnosis of C. auris infections in clinics.
“JP2021122236A” [35] refers to a screening medium containing an enzyme substrate contains raffinose and xylose. C. auris nonassimilated carbon sources as xylose and assimilated raffinose as carbon sources[53].
Conclusions and Perspectives
The whole process of discovering and developing new drugs and diagnostic methods are extensive and expensive. Launching a new drug into the pharmaceutical market can take about 13 years and costs around two billion US dollar. Deeper insights into fungal proteomics, genomics, enzymology, key factors for fungal survival and virulence, and molecular mechanism of resistance would definitely aid in development of better drugs and vaccines against these pathogens. The funds, technical and regulatory hassles could be overcome by concerted efforts in this direction. It’s highly desirable to concentrate on developing newer reliable and rapid tests to diagnose and monitor fungal infections, especially against emerging fungal pathogens as C. auris. The vaccines development is urgent and necessary. In addition, proper microbiological identification, rigorous epidemiological surveillance, adequate treatment and prevention and containment strategies, combined with higher awareness on the side of physicians, microbiologists and healthcare workers, are indispensable to limit further spreading of this pathogen.
References
Satoh K, et al. Candida auris sp. Nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol. 2009;53(1):41–4.
Forsberg K, et al. Candida auris: the recent emergence of a multidrug-resistant fungal pathogen. Med Mycol. 2018;57(1):1–12.
Ahmad S, Alfouzan W. Candida auris: epidemiology, diagnosis, pathogenesis, antifungal susceptibility, and infection control measures to combat the spread of infections in healthcare facilities. Microorganisms. 2021;9(4):807.
Rossato L. COVID-19 and CANDIDA AURIS co-infection: an increasing threat. J Trop Pathol. 2021;50(1):73–5.
Chakrabarti A, Sood P. On the emergence, spread and resistance of Candida auris: host, pathogen and environmental tipping points. J Med Microbiol. 2021. https://doi.org/10.1099/jmm.0.001318.
Kordalewska M, Perlin DS. Identification of drug resistant candida auris. Front Microbiol. 2019;10:1918–1918.
Enoch DA, et al. The changing epidemiology of invasive fungal infections. Methods Mol Biol. 2017;1508:17–65.
Lockhart SR, et al. Candida auris for the clinical microbiology laboratory: not your grandfather’s Candida species. Clin Microbiol Newsl. 2017;39(13):99–103.
Lara-Aguilar V, et al. Adaptation of the emerging pathogenic yeast Candida auris to high caspofungin concentrations correlates with cell wall changes. Virulence. 2021;12(1):1400–17.
Rossato L, Colombo AL. Candida auris: what have we learned about its mechanisms of pathogenicity? Front Microbiol. 2018. https://doi.org/10.3389/fmicb.2018.03081.
Mulet Bayona JV, et al. Impact of the SARS-CoV-2 pandemic in Candidaemia, invasive aspergillosis and antifungal consumption in a tertiary hospital. J Fungi. 2021;7(6):440.
WIPO. World Intellectual Property Report 2019: The geography of innovation: Local hotspots, global networks; 2019 [cited 2022; Geneva: World Intellectual Property Organization.
Siegel DS, Guerrero M. The Impact of quarantines, lockdowns, and ‘reopenings’ on the commercialization of science: micro and macro issues. J Manage Stud. 2021;58(5):1389–94.
Lupkin, S. Coronavirus pandemic brings hundreds of U.S. Clinical Trials to a Halt; 2020.
WHO. WHO reveals leading causes of death and disability worldwide: 2000–2019; 2020.
Williams HL. How do patents affect research investments? Ann Rev Econ. 2017;9:441–69.
Xie Y, Zhang C, Lai Q. China’s rise as a major contributor to science and technology. Proc Natl Acad Sci. 2014;111(26):9437–42.
A, A.G.D.,B.S. Andrew, Antifungal agents, like Ibrexafungerp for Candida auris decolonization, S. INC, editors. 2020; US.
K, P.T.F.W.N.P.N.L. Composition and method for treating candida auris infection, F.T.C.C.L. [JP], editors. 2021; US.
Bradley, B. Compositions and methods for the control and treatment of Candida auris, G.H.S. LLC, editors. 2020; US.
[US], S.P.M., P.M.L. [US], Method to treat antimicrobial resistant candida, H.L.S.L. [US], editors. 2020; US.
Robert, D., R. Bruce, Methods and pharmaceutical compositions for treating Candida auris in blood, C. INC, editors 2020; China.
Carl, G., B. Chad, S. Paul. Methods for treating fungal infections, G. CARL, B.C. S, S.P. B, editors. 2018: US.
E, E.J.J., S. Shakti, I.A. S. Methods of treatment for Candida auris infections, L.A.B.R.I.H.U.M. CT, Editor. 2021; US.
Russell, P.E.J.L.D.H.M.W. Compositions and systems for disinfection; 2021.
H, F.H.E., H.A. T, S. Jingtao, Compositions and methods for detection of candida auris, H.L.R. [CH], R.D.G. [DE], R.M.S.I. [US], Editors. 2020;US.
Xiuying, Z., Z. YAN, C. Yanjing, LAMP primer set, kit and detection method of candida auris, B.T.C.G. HOSPITAL, Editor. China.
Meng, X., G. JIE, H. Jingjing, Method for identifying candida auris by high-resolution-ratio melting curve, P.U.M.C.H. CAMS, Editor. 2019; China.
Sara, S.F., P.B. Luis, T.M.M. Angeles, Method for rapid detection of Candida auris and diagnosis of infection caused by this pathogen F.P.L.I.D.H.U.L.F. [ES], U.V. [ES], U.V.P. [ES], Editors. 2020; Spain.
[US], M.B.J., S.J.L. [US], and C.B.N. [US], NMR methods and systems for the rapid detection of candida species, T.B.I. [US], Editor. 2020; US.
[US], S.W., Novel technology to identify Candida auris, S.F.B.L. [US], Editor. 2020; US.
Xiaodong, L., et al., Nucleic acid reagent, kit, system and method for detecting invasive fungi B.A.B.T.C. LTD, Editor. 2019; China
Koichi, M., Y. Mikachi, A.M. Mahdi, Primer set for detecting candida auris, candida auris detection kit, and method for detecting candida auris, U. Teikyo, Editor. 2020; China
LI, H., T. Shuguang, C. Yong, Real-time fluorescence quantitative PCR detection kit for candida auris, special primers for PCR detection, and TaqMan probe, C.P.C.F.D.C. Prevention, Editor. 2020.
Teikyo, U., Screening medium containing an enzyme substrate contains raffinose and xylose., JP2021122236A, Editor. 2021: Japan.
Ghannoum M, et al. Ibrexafungerp: a novel oral triterpenoid antifungal in development for the treatment of candida auris infections. Antibiotics. 2020;9(9):539.
Jallow S, Govender NP. Ibrexafungerp: a first-in-class oral triterpenoid glucan synthase inhibitor. J Fungi. 2021;7(3):163.
Ghannoum M, et al. Efficacy of ibrexafungerp (SCY-078) against Candida auris in an in vivo guinea pig cutaneous infection model. Antimicrob Agents Chemother. 2020. https://doi.org/10.1128/AAC.00854-20.
Mroczyńska M, Brillowska-Dąbrowska A. Review on current status of echinocandins use. Antibiotics. 2020;9(5):227.
Gamal A, et al. Ibrexafungerp, a novel oral triterpenoid antifungal in development: overview of antifungal activity against candida glabrata. Front Cell Infect Microbiol. 2021. https://doi.org/10.3389/fcimb.2021.642358.
Wiederhold NP. Review of T-2307, an investigational agent that causes collapse of fungal mitochondrial membrane potential. J Fungi. 2021;7(2):130.
Kean R, et al. The comparative efficacy of antiseptics against Candida auris biofilms. Int J Antimicrob Agents. 2018;52(5):673–7.
Seleem D, et al. In vitro evaluation of antifungal activity of monolaurin against Candida albicans biofilms. PeerJ. 2016;4:e2148–e2148.
Liu Y, et al. Taurolidine lock solutions for the prevention of catheter-related bloodstream infections: a systematic review and meta-analysis of randomized controlled trials. PLoS ONE. 2013;8(11):e79417–e79417.
Billamboz M, et al. Promising drug candidates and new strategies for fighting against the emerging superbug Candida auris. Microorganisms. 2021;9(3):634.
Singh S, et al. The NDV-3A vaccine protects mice from multidrug resistant Candida auris infection. PLoS Pathog. 2019;15(8):e1007460–e1007460.
Kean R, et al. Surface disinfection challenges for Candida auris: an in-vitro study. J Hosp Infect. 2018;98(4):433–6.
Fasciana T, et al. Candida auris: an overview of how to screen, detect, test and control this emerging pathogen. Antibiotics. 2020;9(11):778.
Yamamoto M, et al. Rapid detection of candida auris based on loop-mediated isothermal amplification (LAMP). J Clin Microbiol. 2018;56(9):e00591-e618.
Kordalewska M, et al. Rapid and accurate molecular identification of the emerging multidrug-resistant pathogen Candida auris. J Clin Microbiol. 2017;55(8):2445–52.
Pla L, et al. Oligonucleotide-capped nanoporous anodic alumina biosensor as diagnostic tool for rapid and accurate detection of Candida auris in clinical samples. Emerg Microb Infect. 2021;10(1):407–15.
Sexton DJ, et al. Evaluation of a new T2 Magnetic Resonance assay for rapid detection of emergent fungal pathogen Candida auris on clinical skin swab samples. Mycoses. 2018;61(10):786–90.
Osei Sekyere J. Candida auris: a systematic review and meta-analysis of current updates on an emerging multidrug-resistant pathogen. MicrobiologyOpen. 2018;7(4):e00578–e00578.
Funding
The authors have not disclosed any funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have not disclosed any competing interests.
Additional information
Handling Editor: Ferry Hagen.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rossato, L., Simionatto, S., Serafini, M.R. et al. New Technologies to Diagnose and Treat a Multidrug-Resistant Candida auris: A Patent Review. Mycopathologia 187, 535–546 (2022). https://doi.org/10.1007/s11046-022-00669-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11046-022-00669-y