Abstract
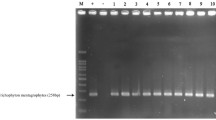
Dermatophytes are among the most successful fungal pathogens in humans, but their virulence mechanisms have not yet been fully characterized. Dermatophytic fungi secrete proteases in vivo, which are responsible for fungal colonization and degradation of the keratinized tissue during infection. In the present study, we used PCR to investigate the presence of genes encoding fungalysins (MEP) and subtilisins (SUB) in three dermatophyte species whose incidence is increasing in Europe: the anthropophilic Trichophyton rubrum (n = 58), zoophilic Microsporum canis (n = 33), and Trichophyton benhamiae (n = 6). MEP2 and SUB4 genes were significantly correlated with T. rubrum; MEP3 and SUB1 were mostly frequently harbored by M. canis; and MEP1, 2, and 4 and SUB3–7 were most frequently harbored by T. benhamiae isolates (p < 0.05). Furthermore, MEP1–5 and SUB1–3 genes were significantly more prevalent among human clinical isolates of M. canis (n = 17) than among asymptomatic cat isolates of M. canis (n = 16; p < 0.05). Unidentified MEP and/or SUB genes in some isolates in the current study may suggest that other gene repertoires may be involved in the degradation of keratin. The presented analysis of the incidence of MEP and SUB virulence genes in three dermatophyte species of diverse origins provides an insight into the host–fungus interaction and dermatophyte pathogenesis.


Similar content being viewed by others

References
Verma S, Madhu R. The great Indian epidemic of superficial dermatophytosis: An appraisal. Indian J Dermatol. 2017;62(3):227–36.
Ginter-Hanselmayer G, Weger W, Ilkit M, Smolle J. Epidemiology of tinea capitis in Europe: current state and changing patterns. Mycoses. 2007;50(suppl. 2):6–13.
Havlickova B, Czaika VA, Friedrich M. Epidemiological trends in skin mycoses worldwide. Mycoses. 2008;51(suppl. 4):2–15.
Zhan P, Liu W. The changing face of dermatophytic infections worldwide. Mycopathologia. 2017;182(1–2):77–86.
Ilkit M, Durdu M. Tinea pedis: the etiology and global epidemiology of a common fungal infection. Crit Rev Microbiol. 2015;41(3):374–88.
Gräser Y, Monod M, Bouchara JP, et al. New insights in dermatophyte research. Med Mycol. 2018;56(suppl. 1):2–9.
Mercer DK, Stewart CS. Keratin hydrolysis by dermatophytes. Med Mycol. 2019;57(1):13–22.
Martinez DA, Oliver BG, Graser Y, et al. Comparative genome analysis of Trichophyton rubrum and related dermatophytes reveals candidate genes involved in infection. mBio. 2012;3(5):e00259–12.
Petrucelli MF, Peronni K, Sanches PR, et al. Dual RNA-seq analysis of Trichophyton rubrum and HaCat keratinocyte co-culture highlights important genes for fungal-host interaction. Genes (Basel). 2018;9(7):pii:E362.
Monod M. Secreted proteases from dermatophytes. Mycopathologia. 2008;166(5–6):285–94.
Monod M, Capoccia S, Léchenne B, Zaugg C, Holdom M, Jousson O. Secreted proteases from pathogenic fungi. Int J Med Microbiol. 2002;292(5–6):405–19.
Lemsaddek A, Chambel L, Tenreiro R. Incidence of fungalysin and subtilisin virulence genes in dermatophytes. In: Current Research, Technology and Education Topics in Applied Microbiology and Microbial Biotechnology, A. Mendéz-Vilas (ed.), Microbiology Book Series 2, Formatex, Badajoz, Spain. 2010:658–65.
Jousson O, Léchenne B, Bontems O, et al. Multiplication of an ancestral gene encoding secreted fungalysin preceded species differentiation in the dermatophytes Trichophyton and Microsporum. Microbiology. 2004;150(Pt 2):301–10.
Nenoff P, Krüger C, Ginter-Hanselmayer G, Tietz HJ. Mycology—an update. Part 1: Dermatomycoses: causative agents, epidemiology and pathogenesis. J Dtsch Dermatol Ges. 2014;12(3):188–209.
de Hoog GS, Dukik K, Monod M, et al. Toward a novel multilocus phylogenetic taxonomy for the dermatophytes. Mycopathologia. 2017;182(1–2):5–31.
Brouta F, Descamps F, Monod M, Vermout S, Losson B, Mignon B. Secreted metalloprotease gene family of Microsporum canis. Infect Immun. 2002;70(10):5676–83.
Jousson O, Léchenne B, Bontems O, et al. Secreted subtilisin gene family in Trichophyton rubrum. Gene. 2004;339:79–88.
Tarabees R, Sabry M, Abdeen E. Incidence of fungalysins virulence genes (MEP1–5) isolated from infected cases in Egypt. Int J Microbiol Res. 2013;4(2):180–7.
Burmester A, Shelest E, Glöckner G, et al. Comparative and functional genomics provide insights into the pathogenicity of dermatophytic fungi. Genome Biol. 2011;12(1):R7.
Staib P, Zaugg C, Mignon B, et al. Differential gene expression in the pathogenic dermatophyte Arthroderma benhamiae in vitro versus during infection. Microbiology. 2010;156(Pt 3):884–95.
Zhang X, Wang Y, Chi W, et al. Metalloprotease genes of Trichophyton mentagrophytes are important for pathogenicity. Med Mycol. 2014;52(1):36–45.
Zhang H, Rokas A, Slot JC. Two different secondary metabolism gene clusters occupied the same ancestral locus in fungal dermatophytes of the Arthrodermataceae. PLoS ONE. 2012;7(7):e41903.
Turin L, Riva F, Galbiati G, Cainelli T. Fast, simple and highly sensitive double-rounded polymerase chain reaction assay to detect medically relevant fungi in dermatological specimens. Eur J Clin Invest. 2000;30(6):511–8.
Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden T. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134.
McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucl Acids Res. 2004;32(Web Server issue):W20–5.
Robati AK, Khalili M, Hazaveh JSH, Bayat M. Assessment of the subtilisin genes in Trichophyton rubrum and Microsporum canis from dermatophytosis. Comp Clin Pathol. 2018;27(5):1343–7.
Descamps F, Brouta F, Monod M, et al. Isolation of a Microsporum canis gene family encoding three subtilisin-like proteases expressed in vivo. J Invest Dermatol. 2002;119(4):830–5.
Brasch J, Beck-Jendroschek V, Voss K, Uhrlaβ S, Nenoff P. Arthroderma benhamiae strains in Germany: Morphological and physiological characteristics of the anamorphs. Hautarzt. 2016;67(9):700–5.
Peres NT, Sanches PR, Falcão JP, et al. Transcriptional profiling reveals the expression of novel genes in response to various stimuli in the human dermatophyte Trichophyton rubrum. BMC Microbiol. 2010;10:39.
Wang L, Ma L, Leng W, et al. Analysis of the dermatophyte Trichophyton rubrum expressed sequence tags. BMC Genomics. 2006;7:255.
Tran VD, De Coi N, Feuermann M, et al. RNA Sequencing-based genome reannotation of the dermatophyte Arthroderma benhamiae and characterization of its secretome and whole gene expression profile during infection. mSystems. 2016;1(4):e00036–16.
Shi Y, Niu Q, Yu X, et al. Assessment of the function of SUB6 in the pathogenic dermatophyte Trichophyton mentagrophytes. Med Mycol. 2016;54(1):59–71.
Bitencourt TA, Macedo C, Franco ME, et al. Transcription profile of Trichophyton rubrum conidia grown on keratin reveals the induction of an adhesin-like protein gene with a tandem repeat pattern. BMC Genomics. 2016;17:249.
Acknowledgements
We wish to thank Professor Murat Durdu for his expert comments on preliminary draft of the manuscript.
Funding
This study was funded by the Research Fund of Mersin University (project no. 2015-AP3-1230).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Handling Editor: Vishnu Chaturvedi.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kaplan, E., Gonca, S., Kandemir, H. et al. Genes Encoding Proteolytic Enzymes Fungalysin and Subtilisin in Dermatophytes of Human and Animal Origin: A Comparative Study. Mycopathologia 185, 137–144 (2020). https://doi.org/10.1007/s11046-019-00367-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11046-019-00367-2