Abstract
Objective
To seek a rapid and reliable molecular biology method to identify the common pathogenic dermatophyte fungi from clinical samples.
Method
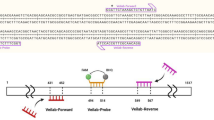
The genome DNA was extracted from cultured strains of seven common dermatophyte fungi species and part of each positive clinical specimen by microscopy. Intergenic spacer regions of ribosomal DNA (ITS) were amplified by semi-nested PCR (snPCR) with three universal primers (NS5, ITS1, and ITS4) for fungi. The amplified products were digested with two restriction endonucleases (BciT130 I, Dde I), the Restriction Fragment Length Polymorphism(RFLP). The rest of each clinical specimen was cultured in Sabouraud’s Agar medium. Then the results of RFLP were compared with the traditional culture results.
Results
The digestion of seven common dermatophyte fungi produced seven different restriction profiles. Restriction profiles of 17 clinical specimens matched, respectively, to that of the cultured strains, and 14 profiles of the 17 ones matched the culture result completely. The coincidence was 100.0%.
Conclusions
snPCR-RFLP analysis of intergenic spacer regions of ribosomal DNA is a valuable method of exactness and clarity for species identification of common dermatophyte fungi from clinical specimens.











Similar content being viewed by others
References
Harmsen D, Schwinn A, Bröcker EB et al. Molecular differentiation of dermatophyte fungi. Mycoses 1999;42:67–70.
Jackson J, Barton C, Evans E, et al. Species identification and strain differentiation of dermatophyte fungi by analysis of ribosomal-DNA intergenic spacer regions. J Clin Microbiol 1999;37:931–6.
Kong X, Yang G, Jin L et al. Molecular determination of dermatophyte fungi using arbitrarily primed polymerase chain reaction. Chin J Dermatol 2001;34:352–4.
Sun X, Yang G, Li W et al. Rapid identification of pathogenic fungi causing Onychomycosis from nial tissue using arbitrarily primed polymerase chain reaction. Clin J Mycol 2007;April 2(2):89.
Xiliang C. Medical molecular biology. People’s Medical Publishing House, 2003; p.186.
Pierre C, Lecossier D, Boussougant Y et al. Use of reamplification protein of Mycobacterium tuberculosis in clinical samples by amplification of DNA. J Clin Microbiol 1991;29:712.
Guo J, Tang Y, Zhang T et al. Detection of Mycoplasma pneumonia by nested polymerase chain reaction. J Zhongshan Medical University 1997;18:229–31.
Stephane C, Claudine S, Samia H et al. Detection of cryptosporidium and identification to the species level by nested PCR and restriction fragment length polymorphism. J Clin Microbiol 2005;43:1017–23.
Makimura K, Tamura Y, Mochizuki T et al. Phylogenetic classification and species identification of dermatophyte strains based on DNA sequences of nuclear ribosomal internal transcribed spacer 1 regions. J Clin Microbiol 1999;37:920–4.
Wang Q, Yi Z, Li H et al. Homology analysis of 18S–28SrDNA of common dermatophytes. Chin J Microbiol Immunol 2006;26:365–8.
Shin JH, Sung JH, Park SJ et al. Species identification and strain differentiation of dermatophyte fungi using polymerase chain reaction amplification and restriction enzyme analysis. J Am Acad Dermatol 2003 Jun;48(6):857–65.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yang, G., Zhang, M., Li, W. et al. Direct Species Identification of Common Pathogenic Dermatophyte Fungi in Clinical Specimens by Semi-nested PCR and Restriction Fragment Length Polymorphism. Mycopathologia 166, 203–208 (2008). https://doi.org/10.1007/s11046-008-9130-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11046-008-9130-3