Abstract
In recent years, there has been a remarkable surge in the development of Natural Language Processing (NLP) models, particularly in the realm of Named Entity Recognition (NER). Models such as BERT have demonstrated exceptional performance, leveraging annotated corpora for accurate entity identification. However, the question arises: Can newer Large Language Models (LLMs) like GPT be utilized without the need for extensive annotation, thereby enabling direct entity extraction? In this study, we explore this issue, comparing the efficacy of fine-tuning techniques with prompting methods to elucidate the potential of GPT in the identification of medical entities within Spanish electronic health records (EHR). This study utilized a dataset of Spanish EHRs related to breast cancer and implemented both a traditional NER method using BERT, and a contemporary approach that combines few shot learning and integration of external knowledge, driven by LLMs using GPT, to structure the data. The analysis involved a comprehensive pipeline that included these methods. Key performance metrics, such as precision, recall, and F-score, were used to evaluate the effectiveness of each method. This comparative approach aimed to highlight the strengths and limitations of each method in the context of structuring Spanish EHRs efficiently and accurately.The comparative analysis undertaken in this article demonstrates that both the traditional BERT-based NER method and the few-shot LLM-driven approach, augmented with external knowledge, provide comparable levels of precision in metrics such as precision, recall, and F score when applied to Spanish EHR. Contrary to expectations, the LLM-driven approach, which necessitates minimal data annotation, performs on par with BERT’s capability to discern complex medical terminologies and contextual nuances within the EHRs. The results of this study highlight a notable advance in the field of NER for Spanish EHRs, with the few shot approach driven by LLM, enhanced by external knowledge, slightly edging out the traditional BERT-based method in overall effectiveness. GPT’s superiority in F-score and its minimal reliance on extensive data annotation underscore its potential in medical data processing.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Cancer continues to be a pressing global public health concern, ranking as the second most prevalent cause of death worldwide. In particular, breast cancer remains the main type of cancer that affects women around the world, achieving the second position in the overall cancer landscape [1,2,3]. According to the World Health OrganizationFootnote 1, the global count of women diagnosed with breast cancer in 2022 reached a staggering 2.26 million. Additionally, cancer treatment continues to be a financially burdensome process that takes a significant social and economic toll on patients and healthcare systems alike.
Integrating EHRs is a promising resource to support clinical research [4]. The cancer care process generates an extensive repository of data that details the progression of cancer within patients [5]. Physicians document these data in EHR systems through narrative-style clinical notes [6]. Extracting and analyzing this information plays a crucial role in supporting oncology research endeavors and improving patient outcomes. However, clinical narratives are written in natural language, which poses a significant challenge in extracting information from these data.
In recent years, the effectiveness of deep learning-driven methodologies has been prominently demonstrated in the extraction of information from clinical narratives [7, 8], including the cancer domain [9, 10]. In the Spanish language, multiple studies have undertaken the task of extracting information from clinical narratives on cancer [11]. However, these efforts have focused mainly on the extraction of medical entities and the identification of negation and uncertainty [12, 13] through separate processes. These studies have represented the extracted information as a NER (Named Entity Recognition) task in which each token in the text is classified into a set of predefined categories [14]. Moreover, these initiatives notably lack a comprehensive methodology for effectively organizing and structuring the extracted information.
The extraction of named entities from unstructured text is of significant importance within the medical domain, facilitating the organization and structuring of patient data. Deep learning-based methodologies employed for entity extraction predominantly operate through two principal approaches: Fine-Tuning and In-context Learning [15]. These techniques contribute to the refinement and optimization of the extraction process, thus enhancing the accuracy and efficiency of information retrieval from medical texts. In what follows, we summarize both approaches:
-
The fine-tuning strategy involves using a pre-trained language model, subjected to further training epochs with annotated data to perform a downstream task [11]. This methodology capitalizes on preexisting language models by adapting them to a designated NER task by incorporating a specifically annotated corpus. This process enhances the model’s performance in a specialized context, optimizing its proficiency in accurately identifying and categorizing named entities within the designated task domain.
-
In-context learning is a method that commonly integrates few-shot demonstrations or prompts during the training process to guide the behavior of the model for a specific task [16]. This approach uses recent advances in Generative Pretrained Transformers (GPT) to address NER tasks, representing a pioneering paradigm, particularly in the intricate realm of processing and structuring complex healthcare data. In-context learning becomes especially relevant when applied to Spanish EHRs, where linguistic nuances pose unique challenges. The transition from traditional NER methods to a GPT-driven approach signifies a crucial shift, underscoring the imperative to assess the efficacy of GPT in comprehending and organizing information within Spanish EHRs.
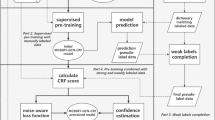
In a previous work [17], a pipeline for structuring breast cancer information from Spanish EHRs using deep learning techniques was developed. The general process is shown in Fig. 1. In this paper we aim to incorporate more functionality of the previous pipeline and in particular we want to analyze GPT as a possible NER generator. For this purpose, in this paper, we compare two strategies for performing clinical NER in the cancer domain: i) a fine-tuning approach using BERT, and ii) an in-context learning method (few shots) using GPT. We examine their strengths and weaknesses, especially in the context of Spanish EHRs. By highlighting the key characteristics, performance metrics, and applicability of each methodology, this study aims to offer a comprehensive understanding of the optimal approach to structuring Spanish EHRs.
Thus, the main contributions of this paper are the following:
-
Comparative Evaluation between BERT and GPT for performing NER: This study presents an in-depth comparative analysis of two NER methodologies applied to Spanish EHRs. The fine-tuning approach using BERT (Bidirectional Encoder Representations from Transformers) is contrasted against the novel method employing GPT. This comparison is crucial to understanding the evolution and effectiveness of NER techniques in the field of healthcare, particularly in the processing of Spanish-language clinical narratives.
-
Analysis of Various Prompting Techniques: In this paper, we have also analyzed different prompt techniques. Various strategies, including few-shot learning and incorporation of external knowledge, are examined. The findings aim to illustrate how skillful prompt use can positively impact the overall performance of NER tasks.
-
Performance and Applicability Evaluation: The paper also aims to identify the most suitable NER approach for Spanish EHRs, considering factors such as processing speed, corpus pre-annotation, model size, transfer learning, or adaptability to diverse medical terminologies. Additionally, research explores the computational efficiency of these NER methods, providing insight into their practicality for real-time applications in healthcare settings. This comprehensive evaluation is essential to make informed decisions about the deployment of AI-driven technologies in resource-constrained environments.
The rest of the paper is organized as follows. Section 2 shows a review of relevant studies on the extraction of cancer concepts, Section 3 presents the problem statement, and Section 4 describes the data and techniques used in this article. Section 5 provides an overview of the experimental results. Subsequently, Section 6 delves into a discussion of these results, while Section 7 outlines the main conclusions and outlook for future work.
2 Related work
The use of deep learning-based methods to extract information from clinical texts has increased in recent years [8, 18]. The main advantages of deep learning are the ability to automatically learn high-level features of texts and the use of transfer learning, where knowledge learned from a previous task is reused to perform a new related task [13, 19]. In the medical domain, Clinical Named Entity Recognition (Clinical NER) aims to extract medical concepts from clinical narratives [14, 20, 21]. Deep learning-based proposals to perform Clinical NER operate through two main approaches: Fine-Tuning and In-Context Learning.
2.1 Fine-tuning
These studies combined pre-trained language models and specific annotated corpora to perform entity extraction as a sequence-labeling task in which each token in the text is labeled with a specific category defined in a corpus [18, 21, 22].
One of the first studies to perform clinical NER using a fine-tuning approach is described in [23]. This study aims to extract lung cancer stages, histology, tumor grades, and therapies (chemotherapy, radiation therapy, surgery) using convolutional neural networks (CNN). The authors highlight the feasibility of extracting cancer-related information from clinical narratives using deep learning methods. In [24] the authors detail a method to extract data from EHRs using machine learning. The technique involves the application of Long Short-Term Memory (LSTM) networks. In [22], the authors proposed a Bidirectional Long Short Memory (BiLSTM) neural network to extract radiotherapy treatments from clinical narratives written in English. This study extracts detailed information related to radiotherapy treatment, such as dose, frequency, fraction frequency, and treatment site. In [25], the authors describe a BiLSTM-based model for the extraction of clinical concepts from German oncological clinical notes. In [9], the authors described a deep learning approach for extracting concepts of breast cancer using BERT. The purpose of this proposal is to extract a comprehensive set of concepts on breast cancer from clinical notes written in Chinese. The authors demonstrate that the BERT-based model outperforms traditional machine learning algorithms to extract named entities in the cancer field. This model supports the extraction of several concepts, such as diagnosis, treatments, and medications. In our publication [17], we introduced an innovative technique for organizing breast cancer data derived from Spanish clinical narratives. This method involved an automated system that employs deep learning to transform these documents into well-structured JSON files for individual patients. The system integrates the extraction of clinical entities and the identification of negation and uncertainty, also using the BERT model for these processes.
2.2 In-context learning
These approaches harness recent advancements in GPT to perform named entity extraction using few-shot demonstrations or prompts during the training process [16]. The arrival of LLMs such as GPT [26] represents a significant step forward in the medical domain. These models could improve the extraction of clinical information through two primary techniques. First, NER, where GPT’s sophisticated understanding of context aids in accurately identifying and categorizing key medical terms and patient data from unstructured narratives. Second, the question-and-answer capability of GPT models offers an innovative approach to information extraction, allowing intuitive natural language querying of complex clinical records [27,28,29].
There are several techniques to extract named entities using the In-context learning approach [30, 31], as follows:
-
Zero-Shot Learning: GPT’s zero-shot approach to NER is especially groundbreaking. In this approach, the model does not need any training on annotated data. Instead, you can use a specific prompt to direct GPT to extract relevant information from a given text and convert it to the desired format. This method is highly efficient and accurate, saving time and resources.
-
Few-Shot Learning: This technique involves providing a few examples to the GPT model to guide it on the task. For instance, if you need to extract specific information, such as cancer type, from a text, you start by giving the model a few examples of cancer type extraction. The model then uses these examples as a reference to perform the task on the new input data. This method is particularly effective because it does not require a lengthy training process with annotated data, which can be time-consuming and resource-intensive.
Recently, several proposals have explored the effectiveness of LLMs in Few-Shot NER [32, 33], as well as in Zero-shot [34]. In [32] GPT-NER is proposed, a novel approach that improves in-context learning performance employing entity-level embedding and self-verification. This approach includes three steps to achieve entity extraction: Task description, Few-shot demonstrations, and input sentence. PromptNER [33], introduces the concept of a chain of thought into NER. This approach leverages the power of Chain-of-Thought Prompting to provide an accessible way to perform a few-shot NER. Prompt-NER requires a list entity type definitions and prompts an LLM to produce a list of potential entities along with corresponding explanations justifying their compatibility. The authors showed comparative results using a 5-shot learning strategy and using several corpora, including the CoNLL dataset [35]. In [36], the authors proposed a few-show learning approach to perform biomedical entity extraction combining query prompts and knowledge-guided instance augmentation. In this approach, the NER task is formulated as a Query Answering (QA) task, then BioBERT [37] is used as a pre-trained model to formulate a query. The model was evaluated in the NCBI [38] dataset using 5-shot, 20-shot, and 50-shot learning examples, obtaining competitive performance.
Although the above studies have shown promising results using Few-shot learning, they perform entity extraction concentrating on the English language. Information extraction in languages other than English has its own challenges [39]. In the case of the Spanish language, there is a lack of approaches to performing clinical NER, given the limited availability of annotated data. These approaches could have an impact on information extraction in the medical domain. Furthermore, it has yet to be explored to use LLMs to perform and evaluate clinical NER in Spanish.
3 Problem statement
Extracting information from EHRs using machine learning models requires the availability of labeled datasets. These datasets are often limited because annotating clinical narratives can be costly and time consuming [40]. The manual clinical text annotation process requires highly skilled annotators and multiple rounds of annotation, increasing medical data annotation costs. Furthermore, the labeled corpora cannot be publicly released due to restrictions on data privacy and patient security. In this scenario, machine learning approaches to dealing with limited annotated data can be crucial to supporting clinical information extraction.
In particular, extracting information from breast cancer clinical narratives presents significant challenges and opportunities in the field of medical informatics. Breast cancer, as one of the most prevalent cancers worldwide, generates a large amount of clinical data. However, these data are often unstructured and recorded in narrative forms within EHR systems, posing a considerable challenge for effective data extraction and analysis. Complexity is further compounded when dealing with EHRs in the Spanish language, which introduces unique linguistic nuances and challenges.
Therefore, the primary problem addressed in this article is the accurate extraction of relevant clinical named entities from Spanish EHRs, specifically those related to breast cancer. The extraction and subsequent structuring of these data are crucial for several reasons:
-
Clinical Research and Patient Care: Structured data are vital for cancer research, allowing analysis of treatment outcomes, patient demographics, and disease progression. It also plays an important role in improving patient care by providing healthcare professionals with easy access to critical patient information.
-
Linguistic Complexity: The Spanish language, with its diverse dialects and idiomatic expressions, presents unique challenges in understanding and processing clinical narratives. The effectiveness of NER tools in accurately identifying and categorizing medical entities in Spanish EHRs is a critical concern.
-
Technological Advancements: With the advent of advanced NLP models such as BERT and GPT, it is necessary to evaluate and compare the effectiveness of these models in processing Spanish EHR. The choice between a traditional model like BERT, known for its robustness in entity recognition, and a more recent, generative model like GPT, capable of handling complex linguistic constructs, is pivotal.
-
Computational Efficiency: Processing large volumes of EHR data requires methods that are not only accurate, but also computationally efficient to facilitate real-time analysis and application in clinical settings.
4 Data and methods
4.1 Annotation of the clinical corpus
In clinical evaluations, in addition to diagnosis, it is also crucial to annotate various other aspects such as prescribed treatments, surgeries performed, the presence and specific location of metastases, as well as any occurrences of recurrence. An in-house annotated corpus has been generated. Two skilled oncology annotators manually annotated a collection of 550 clinical notes. These annotators were guided by a data scientist who led the annotation process. The annotated corpus was designed to facilitate the extraction of medical entities related to breast cancer. Annotations were generated using the Prodigy toolFootnote 2. The annotation schema applied to the corpus is presented in Table 1, with the aim of extracting atomic concepts that improve the structure of the information. In this context, atomic concepts denote clinical entities composed of one or two tokens. Figure 2 illustrates a series of annotations relating to breast cancer. According to this figure, cancer diagnosis, “carcinoma infiltrante de mama derecha tipo nos” is segmented into four medical concepts: Cancer Concept, Cancer Expansion, Cancer Location, and Cancer Type.
4.1.1 Inter-annotation agreement
To evaluate the reliability of expert annotations, we calculated the Inter-annotation agreement (IAA) between two clinical annotators. The IAA measure aims to guarantee similar and consistent annotations. We used the F-measure to calculate the IAA between annotation pairs, as suggested in several NER annotation studies [41,42,43]. The IAA was calculated when oncology experts finished the annotation process, as described in [13]:
-
We considered the first annotator as \(A_1\), and the second annotator as \(A_2\).
-
Any annotation sets (\(A_1\) or \(A_2\)) can be considered the gold standard (correct annotations). Therefore, Precision is the percentage of correct positive annotations made by the other annotator. Recall is the percentage of positive annotations made by the other annotator. The F-measure is the harmonic mean between Precision and Recall, as shown in (4.1). We obtained an F-measure of 90%, which suggests that the annotations are consistent between two annotators.
After measuring the IAA, we perform a disagreement resolution step to improve the reliability of the corpus. The annotators met with the data scientist who led the annotation process and reviewed the cases where there were disagreements. The clinician annotators then resolved the discrepancies guided by the data scientist.
4.2 Information extraction with BERT and GPT
BERT is a transformer-based model known for its deep bidirectional nature. It is pretrained on a large corpus using a Masked Language Model (MLM) and Next Sentence Prediction (NSP), allowing it to capture a rich understanding of language context and semantics. BERT’s architecture enables it to consider the full context of a sentence (both left and right of every word) during training, which is particularly beneficial for tasks that require a deep understanding of language structure such as NER. It is crucial to emphasize that to achieve optimal performance in NER tasks, BERT requires fine-tuning on task-specific datasets. This fine-tuning process is essential for adapting the model to the specific nuances and requirements of the target application.
In contrast, GPT is an autoregressive transformer model that focuses primarily on generative tasks. It uses a left-to-right training approach, where each word is predicted based on the preceding words. Although less effective in understanding the bidirectional context, this method is powerful in generating coherent and contextually relevant text. Furthermore, GPT can be highly competitive in NER tasks when employing few-shot learning techniques and integrating external knowledge, leveraging its advanced generative capabilities to effectively understand and categorize complex data. Given the complexity and time-intensive nature of annotating clinical notes, a few-shot learning techniques become particularly valuable. These methods allow models to efficiently learn and adapt from a limited set of examples, which is ideal in scenarios such as healthcare documentation, where extensive data collection is often impractical or impossible. This approach is especially relevant for handling the intricate details and depth of information contained in medical records.
The choice between BERT and GPT for NER tasks largely depends on the specific requirements of the task and the nature of the dataset. Figure 3 demonstrates how NER techniques can be implemented using both GPT and BERT.
BERT vs. GPT for NER: To evaluate the capabilities of GPT and BERT in a comparable manner, particularly in terms of extracting entities from sentences, a distinct approach must be employed for each model. For example, when processing the sentence “Carcinoma ductal de mama”, BERT can identify all entities in a single prediction. However, GPT requires a separate prompt for each entity to achieve the same task. This necessitates the development of specific prompts for GPT to enable a fair comparison of entity extraction performance between the two models
4.2.1 Fine-tunning for BERT
This step aims to extract breast cancer concepts from clinical narratives using a BERT style deep learning-based model. The model performs medical concept extraction as a sequence-labeling task where each token of a text sentence is classified with its corresponding label in the necessary cases. This step is divided into two subtasks as follows:
-
1.
Corpus annotation: This phase is explained in Section 4.1, involves the manual or automated labeling of entities in a given text corpus to create a training dataset for the BERT model; a sample is depicted in Fig. 2.
-
2.
Training a model: With the label corpus, we trained a model to extract medical concepts using multilingual BERT [44]. This training approach uses a transfer learning technique to perform clinical NER in the breast cancer field. Transfer learning is achieved by fine-tuning the BERT model with a classification layer on top, as described in [13]. The trained model is capable of performing named entity recognition. The model obtained processes clinical texts at the sentence level. That is, the model inputs a sentence text and outputs a set of entities extracted from the sentence. The model labels the entities using the predefined labels in the annotation scheme described in Table 1.
Prompt Structure: the prompt is divided into four sections: the role task, the format and task description, the few-shot examples, and finally, the task to solve. This order allows the LLM to comprehend the task effectively, as elucidated in [45]
4.2.2 Structure of prompts for GPT models
In the domain of NER for clinical texts using GPT models, a variety of methodologies and strategies have been used, focusing on the design, optimization and refinement of the prompts. A critical aspect of successfully implementing NER with GPT lies in designing a well-defined and strategically structured prompt.
This method uses a separate extraction process for each entity, where unique prompts are made for each one. These prompts share a similar structure but contain specific details or knowledge unique to each entity. Each prompt follows the same format as illustrated in Fig. 4, ensuring consistency in structure between different types of entities.
The fundamental aspects of this process fall into four main categories:
-
Common Prompt Structure: Two variations of prompt models have been designed: one incorporating external knowledge and the other without it. While both maintain the same structure, they differ primarily in the extent to which external knowledge is embedded within them. This structure includes:
-
Context: The task to be performed by the model is defined, specifying the particular entity to be extracted and the extraction method.
-
Information: The model is provided with data about the entity to be extracted, highlighting the required format.
-
Instructions: Detailed instructions on the process to follow are presented. Clarity in explaining the format required for entity extraction is crucial, as is a detailed description of how entities are typically presented.
-
-
Two Types of Prompts:
-
Prompt without External Knowledge: provides the model with little additional information, being limited mainly to the entity name. Its objective is to evaluate the model’s ability to operate with minimal information and compare the results with those obtained with more detailed prompts.
-
Prompt with External Knowledge: Provides a greater amount of information and knowledge about the entity, which improves the contextualization and understanding of the task. Our external knowledge is made up of concept_words, where the most common vocabulary related to the entity we are exploring is included. This helps refine our search and analysis, ensuring that responses are more aligned with the specific domain of the entity.
-
-
Zero and Few-shot Strategies: These techniques have been applied for both prompts models. Inclusion of specific examples has resulted in increased precision. In the few-shot approach, we have utilized five examples (5-shot), selected entirely at random from the pool of notes that contain at least one label related to the specified entity.
-
Formatting of Entities: As described in [32], a specific format has been defined for the output text, which consists of keeping the original text except for the entity of interest, which must be delimited between @@ and ##. Thus, the entity is marked in its original position in the text as @@ entity ##. This format has been used to facilitate the extraction and validation of the entities, as well as to calculate the final metrics.
4.3 Computational environment
The experiments were programmed in Python 3.10, and the source code is available at: https://github.com/Alvaro8gb/BERTvsGPT.
For training the BERT model, we utilized an NVIDIA GeForce RTX 3090, operating on a Linux system with kernel version 5.15.0-83-generic, paired with an Intel Core i9-7900X CPU, based on Ubuntu 20.04.1 LTS.
To take advantage of the GPT models, we utilize the OpenAI APIFootnote 3 through the Azure Portal provided by Microsoft.
5 Experimentation and results
5.1 Metrics
To evaluate the performance of the proposed approach, we used the following standard metrics. Precision (5.1), Recall (5.2), and F-score (5.3). The F-score is calculated as a weighted average of the Precision and Recall measurements. A clinical entity is correctly classified when the predicted label is equal to the label indicated by the annotated corpus. The performance of the medical concept extraction task is measured at the entity level.
5.2 Validation
To evaluate the described BERT model, we used a k-fold cross-validation strategy with \(k = 10\). The model performance was computed as the average across all ten-folds executed during the cross-validation process. This BERT model was trained using PyTorchFootnote 4 and SpaCy toolsFootnote 5. The hyperparameters used included a batch size of 256, a dropout rate of 0.1, a total of 10 epochs, a learning rate of 0.001, and the optimizer used was “Adam”.
To evaluate GPT models, all instances are utilized for evaluation except those employed in few-shot techniques, as these models do not require additional training. The selected temperature for the GPT model in the experiments is set to 0, meaning that the model will choose the token with the highest probability. This choice increases the confidence and determinism of the model’s predictions in the output.
5.3 Results
Table 2 presents a comparative analysis of performance between two versions of the OpenAI language models, GPT-3.5 and GPT-4, using different prompt engineering techniques. Data are organized into columns representing prompt type, precision, recall, and F-Score for each model in various configurations.
In particular, for the GPT-3.5 model, the following has been obtained:
-
When using an approach with 5 examples plus external knowledge (5-shot + ext.), the model achieves a precision of 0.72, a recall of 0.57, and an F-Score of 0.64.
-
With just 5 examples, precision is at 0.67, recall at 0.54, and F-Score at 0.60.
-
In a zero-example approach with external knowledge (Zero-shot + ext.), the values are significantly lower: 0.40 for precision, 0.17 for recall, and 0.24 for F-Score.
-
Without examples or external knowledge (Zero-shot), the values are the lowest with a precision of 0.06, a recall of 0.05, and an F-Score of 0.05.
On the other hand, when we use the GPT-4 model these results are obtained:
-
With 5 examples and external knowledge (5-shot + ext.), there are notable improvements with a precision of 0.92, a recall of 0.97, and an F-Score of 0.95.
-
Using only five examples (5 shots), precision is 0.85, recall is 0.82, and F-Score is 0.83.
-
With zero examples and external knowledge (Zero-shot + ext.), precision is 0.98, recall at 0.76, and F-Score at 0.86.
-
In the zero-example approach without external knowledge (Zero-shot), the values are 0.24 for precision, 0.20 for recall, and 0.22 for F-Score.
The results displayed in Table 2 clearly show that the GPT-4 model outperforms GPT-3.5 in all configurations and that the inclusion of external knowledge and the use of examples significantly improve the performance of both models. The best F-Score presented is for the GPT-4 model using the few-shot approach with external knowledge (5-shot + ext.), which is 0.95. Additionally, the GPT-4 model with external knowledge and without examples (Zero-shot + ext.) presents an outstanding performance in precision. Therefore, we can observe that a smaller model like GPT-3.5 exhibits higher hallucinations, as its recall is lower compared to the GPT-4 configurations. The highest recall, 0.97, is achieved for the 5-shot scenario with external knowledge.
In Table 3, the global results for the BERT and GPT models in the context of NER for various medical entities are summarized.
For the BERT model, the overall precision across all categories is 0.93, with a recall of 0.94, and an F-score of 0.94. This indicates a high level of accuracy and consistency in the ability of the BERT model to correctly identify and classify entities across a range of cancer-related categories.
In comparison, the GPT model also shows strong performance with an overall precision of 0.92, which is slightly lower than BERT. However, GPT surpasses BERT in recall with a score of 0.97, suggesting that it is more comprehensive in identifying relevant entities. The F-score for GPT, which balances precision and recall, is 0.95, slightly higher than that for BERT.
In the performance of the GPT model, there are 10 values that exceed 0.95 in the F-score metric. This impressive level of performance highlights the GPT’s strong capability for accurately identifying medical entities, which is incredibly helpful for simplifying the data preparation phase in medical research. By greatly reducing the need for extensive initial corpus annotation, GPT provides a significant advantage in efficiency, making the processing of medical text data much more straightforward and effective.
In the context of GPT’s performance on NER, the F-score values that dip below 0.9 can be attributed to the model encountering similar terminology among various entities. While it is feasible to rectify this by fine-tuning the prompts, it should be noted that all prompts were designed to be consistent across different entity types. This standardization was deliberate to ensure uniform testing conditions. Therefore, while prompt optimization could enhance the model’s ability to distinguish between entities with similar terms, such alterations were not implemented in order to preserve the uniform methodology in the evaluation process.
6 Discussion
In this paper, we have presented a comparative analysis of two state-of-the-art language models, BERT and GPT, in the context of NER for medical entities, as depicted in Table 4. Based on the results obtained, it is evident that both models exhibit strong capabilities, with GPT marginally outperforming BERT in overall F-score (0.95 vs. 0.94).
BERT’s requirement for extensive pre-annotation of the corpus signifies a significant investment in manual effort and time. Despite this, its fine-tuning requirement, although a resource-intensive process, allows for a tailored fit to specific NER tasks, which can result in highly accurate model performance. On the other hand, GPT simplifies the workflow by reducing the need for extensive data preparation, although it still requires some expertise to develop these prompts effectively.
BERT demonstrates rapid inference times, typically in the order of milliseconds per entity, rendering it highly suitable for real-time applications. On the contrary, GPT requires substantial computational resources, often reliant on cloud services, which may incur additional costs and dependencies.
Both models exhibit exceptional transfer learning capabilities; however, GPT’s less data-intensive nature and its capacity to adapt to new domains without necessitating retraining render it particularly appealing for applications requiring swift deployment across diverse medical subfields. Moreover, GPT’s high level of customizability and resilience to ambiguity make it a versatile tool, adept at navigating the intricacies of diverse medical terminology and nuanced language commonly encountered in medical records.
Lastly, GPT’s potential for Zero-shot learning presents a paradigm shift, enabling it to predict entities in medical texts without prior fine-tuning on specific NER tasks. This capability positions GPT as a formidable tool for medical NER tasks, facilitating rapid scalability and deployment across diverse medical datasets and domains.
7 Conclusion
In this study, we have compared the BERT and GPT models for the extraction of medical entities using NER tasks. Our analysis reveals that BERT offers slightly better precision and excels in real-time processing tasks. However, GPT stands out with its marginally higher F-score and exceptional adaptability to new domains, requiring little or no additional training, positioning it as a compelling option for diverse applications.
The efficiency of GPT in handling tasks with minimal pre-annotation demonstrates its potential to reduce the time and resources typically required for model training and deployment.
Integration of these models into real-world clinical systems presents fertile ground for future work. There is a need to assess the practicality of implementing such advanced NER systems in diverse healthcare settings and to assess their impact on workflow efficiency and patient outcomes.
In conclusion, while both BERT and GPT show excellent performance in medical entity extraction, their differences in resource requirements, processing times, and flexibility in deployment suggest that the choice between them should be guided by the specific needs and constraints of the use case at hand.
8 Limitations
This paper emphasizes specific constraints. First, employing the OpenAI API incurs a significant cost, rendering it impractical for real-world data science challenges, especially those involving large-scale datasets. Another limitation is the issue of data privacy, as sending medical data to cloud servers may not be possible due to data protection concerns. However, having a local LLM can solve both problems.
Data Availability
Breast cancer corpus is available “on request”. This corpus can be accessible after an evaluation by the hospital’s ethics committee. To request access to anonymized data, please contact Dr. Mariano Provencio at the following email: mariano.provencio@salud.madrid.org. The supporting data associated with this article can be downloaded at https://github.com/Alvaro8gb/BERTvsGPT.
References
Groot PM, Wu CC, Carter BW, Munden RF (2018) The epidemiology of lung cancer. Transl Lung Cancer Res 7(3)
Lung Health and Diseases Lung Disease Lookup. https://www.lung.org/lung-health-and-diseases/lung-disease-lookup/lung-cancer/resource-library/lung-cancer-fact-sheet.html. Accessed 30 Jan 2020
Lung Health and Diseases Lung Disease Lookup. https://www.cancer.org/cancer/lung-cancer/about/key-statistics.html. Accessed 14 Feb 2020
Spasić I, Livsey J, Keane JA, Nenadić G (2014) Text mining of cancer-related information: Review of current status and future directions. Int J Med Inform 83(9):605–623. https://doi.org/10.1016/j.ijmedinf.2014.06.009
Kehl KL, Xu W, Lepisto E, Elmarakeby H, Hassett MJ, Van Allen EM, Johnson BE, Schrag D (2020) Natural language processing to ascertain cancer outcomes from medical oncologist notes. JCO Clin Cancer Inform 4:680–690. https://doi.org/10.1200/cci.20.00020
Bose P, Srinivasan S, Sleeman WC, Palta J, Kapoor R, Ghosh P (2021) A survey on recent named entity recognition and relationship extraction techniques on clinical texts. Appl Sci (Switzerland) 11(18):1. https://doi.org/10.3390/app11188319
Yang X, Zhang H, He X, Bian J, Wu Y et al (2020) Extracting family history of patients from clinical narratives: exploring an end-to-end solution with deep learning models. JMIR Med Inform 8(12):22982
Wu S, Roberts K, Datta S, Du J, Ji Z, Si Y, Soni S, Wang Q, Wei Q, Xiang Y et al (2020) Deep learning in clinical natural language processing: a methodical review. J Am Med Inform Assoc 27(3):457–470
Zhang X, Zhang Y, Zhang Q, Ren Y, Qiu T, Ma J, Sun Q (2019) Extracting comprehensive clinical information for breast cancer using deep learning methods. Int J Med Inform 132(September):103985. https://doi.org/10.1016/j.ijmedinf.2019.103985
Hernandez-Boussard T, Kourdis PD, Seto T, Ferrari M, Blayney DW, Rubin D, Brooks JD (2017) Mining electronic health records to extract patient-centered outcomes following prostate cancer treatment. AMIA ... Annual Symposium proceedings. AMIA Symposium 2017, pp 876–882
Solarte-Pabón O, Torrente M, Garcia-Barragán A, Provencio M, Menasalvas E, Robles V (2022) Deep learning to extract breast cancer diagnosis concepts. In: 2022 IEEE 35th international symposium on computer-based medical systems (CBMS), pp 13–18. https://doi.org/10.1109/CBMS55023.2022.00010
Santiso S, Pérez A, Casillas A, Oronoz M (2020) Neural negated entity recognition in Spanish electronic health records. J Biomed Inform 105(December 2019):103419. https://doi.org/10.1016/j.jbi.2020.103419
Pabón OS, Montenegro O, Torrente M, González AR, Provencio M, Menasalvas E (2022) Negation and uncertainty detection in clinical texts written in spanish: a deep learning-based approach. PeerJ Comput Sci 8:913
Pagad NS, Pradeep N (2022) Clinical named entity recognition methods: an overview. In: International conference on innovative computing and communications: proceedings of ICICC 2021, vol 2, pp 151–165. Springer
Mosbach M, Pimentel T, Ravfogel S, Klakow D, Elazar Y (2023) Few-shot fine-tuning vs. in-context learning: a fair comparison and evaluation. Preprint arXiv:2305.16938
Ge Y, Guo Y, Das S, Al-Garadi MA, Sarker A (2023) Few-shot learning for medical text: a review of advances, trends, and opportunities. J Biomed Inform 104458
García-Barragán A, Solarte-Pabón O, Nedostup G, Provencio M, Menasalvas E, Robles V (2023) Structuring breast cancer spanish electronic health records using deep learning. In: 2023 IEEE 36th international symposium on computer-based medical systems (CBMS), pp 404–409. https://doi.org/10.1109/CBMS58004.2023.00252
Wei Q, Ji Z, Li Z, Du J, Wang J, Xu J, Xiang Y, Tiryaki F, Wu S, Zhang Y et al (2020) A study of deep learning approaches for medication and adverse drug event extraction from clinical text. J Am Med Inform Assoc 27(1):13–21
Harerimana G, Kim JW, Yoo H, Jang B (2019) Deep learning for electronic health records analytics. IEEE Access 7:101245–101259
Fu S, Chen D, He H, Liu S, Moon S, Peterson KJ, Shen F, Wang L, Wang Y, Wen A, Zhao Y, Sohn S, Liu H (2020) Clinical concept extraction: A methodology review. J Biomed Inform 109:103526. https://doi.org/10.1016/j.jbi.2020.103526
Solarte-Pabón O, Montenegro O, García-Barragán A, Torrente M, Provencio M, Menasalvas E, Robles V (2023) Transformers for extracting breast cancer information from spanish clinical narratives. Artif Intell Med 143:102625. https://doi.org/10.1016/j.artmed.2023.102625
Bitterman D, Chen Lin H, Finan S, Warner J, Mak R, Savova G (2020) Extracting radiotherapy treatment details using neural network-based natural language processing. In: Annual Meeting of the American Society for Radiation Oncology, Cham
Wang L, Luo L, Wang Y, Wampfler J, Yang P, Liu H (2019) Natural language processing for populating lung cancer clinical research data. BMC Med Inform Decis Mak 19(Suppl 5):1–10. https://doi.org/10.1186/s12911-019-0931-8
Adamson B, Waskom M, Blarre A, Kelly J, Krismer K, Nemeth S, Gippetti J, Ritten J, Harrison K, Ho G, Linzmayer R, Bansal T, Wilkinson S, Amster G, Estola E, Benedum CM, Fidyk E, Estévez M, Shapiro W (2023) Cohen AB (2023) Approach to machine learning for extraction of real-world data variables from electronic health records. Front Pharmacol 14. https://doi.org/10.3389/fphar.2023.1180962
Kittner M, Lamping M, Rieke DT, Götze J, Bajwa B, Jelas I, Rüter G, Hautow H, Sänger M, Habibi M, Zettwitz M, Bortoli T, Ostermann L, Ševa J, Starlinger J, Kohlbacher O, Malek NP, Keilholz U, Leser U (2021) Annotation and initial evaluation of a large annotated German oncological corpus. JAMIA Open 4(2):1–9. https://doi.org/10.1093/jamiaopen/ooab025
Brown T, Mann B, Ryder N, Subbiah M, Kaplan JD, Dhariwal P, Neelakantan A, Shyam P, Sastry G, Askell A et al (2020) Language models are few-shot learners. Adv Neural Inf Process Syst 33:1877–1901
Nori H, King N, McKinney SM, Carignan D, Horvitz E (2023) Capabilities of GPT-4 on medical challenge problems
Chada R, Natarajan P (2021) Fewshotqa: A simple framework for few-shot learning of question answering tasks using pre-trained text-to-text models, pp 6081–6090. https://doi.org/10.18653/v1/2021.emnlp-main.491
Singhal SAea (2023) Large language models encode clinical knowledge. Nature 620:172–180. https://doi.org/10.1038/s41586-023-06291-2
Labrak Y, Rouvier M, Dufour R (2023) A Zero-shot and Few-shot study of instruction-finetuned large language models applied to clinical and biomedical tasks
Li M, Zhang R (2023) How far is Language Model from 100 Medical Domain
Wang S, Sun X, Li X, Ouyang R, Wu F, Zhang T, Li J, Wang G (2023) GPT-NER: named entity recognition via large language models
Ashok D, Lipton ZC (2023) PromptNER: prompting for named entity recognition
Kaufmann B, Busby D, Das CK, Tillu N, Menon M, Tewari AK, Gorin MA (2024) Validation of a zero-shot learning natural language processing tool to facilitate data abstraction for urologic research. Eur Urol Focus. https://doi.org/10.1016/j.euf.2024.01.009
Sang EF, De Meulder F (2003) Introduction to the conll-2003 shared task: Language-independent named entity recognition. Preprint arXiv:cs/0306050
Chen P, Wang J, Lin H, Zhao D, Yang Z (2023) Few-shot biomedical named entity recognition via knowledge-guided instance generation and prompt contrastive learning. Bioinformatics 39(8):496. https://doi.org/10.1093/bioinformatics/btad496. https://academic.oup.com/bioinformatics/article-pdf/39/8/btad496/51226065/btad496.pdf
Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, Kang J (2019) BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 36(4):1234–1240. https://doi.org/10.1093/bioinformatics/btz682. https://academic.oup.com/bioinformatics/article-pdf/36/4/1234/32527770/btz682.pdf
Doğan RI, Leaman R, Lu Z (2014) Ncbi disease corpus: a resource for disease name recognition and concept normalization. J Biomed Inform 47:1–10
Névéol A, Dalianis H, Velupillai S, Savova G, Zweigenbaum P (2018) Clinical Natural Language Processing in languages other than English: Opportunities and challenges. J Biomed Semant 9(1):1–13. https://doi.org/10.1186/s13326-018-0179-8
Ge Y, Guo Y, Das S, Al-Garadi MA, Sarker A (2023) Few-shot learning for medical text: A review of advances, trends, and opportunities. J Biomed Inform 144:104458. https://doi.org/10.1016/j.jbi.2023.104458
Hripcsak G, Rothschild AS (2005) Agreement, the F-measure, and reliability in information retrieval. J Am Med Inform Assoc 12(3):296–298. https://doi.org/10.1197/jamia.M1733
Dalianis H (2018) Evaluation metrics and evaluation, pp 45–53. Springer, Cham. https://doi.org/10.1007/978-3-319-78503-5_6
Campillos-Llanos L, Valverde-Mateos A, Capllonch-Carrión A, Moreno-Sandoval A (2021) A clinical trials corpus annotated with UMLS entities to enhance the access to evidence-based medicine. BMC Med Inform Decis Mak 21(1):1–19. https://doi.org/10.1186/s12911-021-01395-z
Devlin J, Chang MW, Lee K, Toutanova K (2019) BERT: Pre-training of deep bidirectional transformers for language understanding. NAACL HLT 2019 - 2019 conference of the North American chapter of the association for computational linguistics: human language technologies - proceedings of the conference 1(Mlm), pp 4171–4186. arXiv:1810.04805
Giray L (2023) Prompt engineering with chatgpt: A guide for academic writers. Ann Biomed Eng 1–5
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature. This article is supported by the Horizon 2020 research and innovation program of the European Union under grant agreement No. 875160, CLARIFY project. Additionally, we extend gratitude for the collaboration facilitated by the agreement between the Universidad Politécnica de Madrid (UPM) and Microsoft, which allows for the utilization of Azure credits.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix A: Prompts
Appendix A: Prompts
This section shows some examples of prompts designed to guide GPT models in the task of extracting entities from clinical texts on breast cancer.
Few-shot prompt with external knowledge: example of a prompt with external knowledge with an example for the extraction of cancer concept entity. More information and context about the entity to be extracted is provided. The accuracy of the information provided varies depending on the difficulty the model may encounter in identifying the entity. One-shot learning: an example clinical note is presented to the model to facilitate understanding of the task and the format required for accurate entity extraction
These prompts Figs. 5 and 6, are designed and formulated to take advantage of the potential NER capabilities of GPT models, allowing accurate identification and classification of clinical terms. Each example prompt illustrates the specific interaction between the user and the system for effective extraction of relevant entities for the purpose of studying the adaptation of these pre-trained language models to understand and process specialized terminology within the medical domain.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
García-Barragán, Á., González Calatayud, A., Solarte-Pabón, O. et al. GPT for medical entity recognition in Spanish. Multimed Tools Appl (2024). https://doi.org/10.1007/s11042-024-19209-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11042-024-19209-5