Abstract
The generalisability of deep neural network classifiers is emerging as one of the most important challenges of our time. The recent COVID-19 pandemic led to a surge of deep learning publications that proposed novel models for the detection of COVID-19 from chest x-rays (CXRs). However, despite the many outstanding metrics reported, such models have failed to achieve widespread adoption into clinical settings. The significant risk of real-world generalisation failure has repeatedly been cited as one of the most critical concerns, and is a concern that extends into general medical image modelling. In this study, we propose a new dataset protocol and, using this, perform a thorough cross-dataset evaluation of deep neural networks when trained on a small COVID-19 dataset, comparable to those used extensively in recent literature. This allows us to quantify the degree to which these models can generalise when trained on challenging, limited medical datasets. We also introduce a novel occlusion evaluation to quantify model reliance on shortcut features. Our results indicate that models initialised with ImageNet weights then fine-tuned on small COVID-19 datasets, a standard approach in the literature, facilitate the learning of shortcut features, resulting in unreliable, poorly generalising models. In contrast, pre-training on related CXR imagery can stabilise cross-dataset performance. The CXR pre-trained models demonstrated a significantly smaller generalisation drop and reduced feature dependence outwith the lung region, as indicated by our occlusion test. This paper demonstrates the challenging problem of model generalisation, and the need for further research on developing techniques that will produce reliable, generalisable models when learning with limited datasets.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Effective classification of samples from a limited dataset has been a long-sought goal in the field of computer vision. The problem is particularly pronounced in medical applications, where the availability of high-quality, labelled datasets is naturally constrained by the limited availability of skilled annotators and restrictive legislation regarding data privacy. To overcome the challenge of low volumes of annotated training data in the medical setting, transfer learning has been used extensively in the literature [1]. However, a consequence of the low availability of data is that thorough evaluation of model generalisation capacity is often overlooked in the literature.
Generalisation failure is a well-established challenge in machine learning. One of the earliest studies to highlight this issue was published by Torralba and Efros [2], who demonstrated that large training sets can still produce biased models unable to perform well on new data. Such concerns persist, with recent works revealing the presence of spurious correlations in the popular ImageNet dataset [3], that can cause models to rely on irrelevant and non-generalising decision boundaries [4]. Similarly, recent critical publications of medical image models repeatedly warn that the risk of biased predictions and generalisation failures prevented their adoption into the clinical setting [5, 6]. As such, the evaluation of model generalisation capacity is a critical aspect which must be considered when developing deep learning methods, especially for medical applications. This is exemplified in the COVID-19 medical image model literature.
The COVID-19 pandemic brought global disruption and created a high demand for fast and accurate testing. Hospitals began to incorporate the use of chest x-rays (CXRs) to speed up the triage of suspected COVID-19 patients [7], and this, in turn, led to a sudden rise in the availability of COVID-19 labelled CXR data. However, the identification of COVID-19 abnormalities in CXRs was observed to be a challenging task. COVID-19 indicators can overlap with other viral infections, such as influenza [8], which may lead to an incorrect diagnosis of COVID-19. Furthermore, in mild cases, an x-ray may not capture the abnormalities at a visible level [7]. The authors of [9] reported cases where indicators of COVID-19 infection were present in computed tomography (CT) scans of COVID-19 patients, but not visible in the accompanying CXRs. During the early stages of the pandemic, radiologists struggled to identify all cases using this method, with some reporting that their radiologists achieved 64% sensitivity when trying to detect COVID-19 from CXRs alone [10]. As such, significant potential for improved radiological diagnosis using AI as an assistive technology was established [6].
A surge of research activity emerged to develop technology to assist radiologists in diagnosing COVID-19 from CXRs. A significant portion of this research was focused on the development of AI models to detect and localise COVID-19, with many publications utilising transfer learning in conjunction with existing CXR datasets [1]. One of the earliest and most heavily cited examples was the bespoke DarkCOVIDNet [11], which at the time of writing accrued over 2000 citations. The architecture achieved an outstanding average binary classification accuracy of 98.8% with an average sensitivity of 95.13%, while working with a dataset of 127 COVID-19 images with negative samples taken from [12]. Similarly, using only 358 COVID-19 CXRs, the custom-designed COVID-Net [13] architecture which has gained more than 2400 citations, reported overall accuracy of 93.3% and a COVID-19 sensitivity of 91%. This trend of model training and evaluation using small datasets continued throughout the COVID-19 model publication cycle, however the challenge of evaluating models’ capacity for generalisation remained often overlooked [14].
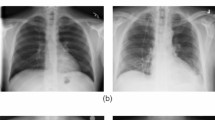
Visual comparison of image samples taken from the internal training dataset, COVID-Xray-5k [18]. Image (a) is a high resolution image from the COVID-19 samples, while image (b) is a significantly lower resolution image from the negative samples
Beyond the broad risk of generalisation failure, medical imagery poses unique challenges due to characteristics of the medium itself. There are growing concerns that models may rely on non-pathological features to distinguish between positive and negative cases. This risk increases when training datasets contain class-wise stratifications irrelevant to the task. As reported by Roberts et al. [6], various COVID-19 papers used the popular composite COVID-19 chest x-ray dataset [15], in conjunction with the Guangzhou Pneumonia dataset [16]. However, the pneumonia samples are exclusively paediatric images, in contrast to the wide age range present in the COVID-19 images [14]. As shown by DeGrave et al. [17], medical deep learning models tend to optimise their performance by relying on the easiest distinguishing features, and in this scenario, this may lead to optimisation based on distinguishing between paediatric and adult images. Moreover, the covid-xray-5k [18] dataset used in this study contains resolution stratification, where all negative samples obtained from the CheXpert dataset have maximal dimensions of 500x500px, whereas the COVID-19 samples range in dimensions from 500 to 3500px. These differences can be seen in Fig. 1, where image (a) shows a very high-resolution image of the chest region, whereas in image (b), the pixels are clearly visible, indicating information loss. Such differences may present confounding factors that could also facilitate shortcut learning [19] and threaten generalisation capacity.
This paper investigates aspects of generalisability within the field of chest x-rays, specific to the classification of COVID-19. We investigate the impact of training with small, imbalanced datasets on model generalisation, and whether transfer learning can significantly improve their ability to accurately predict on unseen data sources. This work is motivated by recent literature that emphasises how uncertainty and concerns regarding the reliability and robustness of medical image models pose a significant barrier to their adoption into clinical settings. We shine light on this problem, by quantifying the extent to which COVID-19 CXR models can generalise to unseen datasets and determining their susceptibility to rely upon shortcut features. We compare a variety of architectures, representing the most commonly used in COVID-19 publications, as well as the state-of-the-art. We conclude our study with a chest occlusion evaluation to identify models heavily dependent on features outside the lung tissue region and determine whether this correlates with poorly generalising models. Our work differs from that of [20] and [21] in that we look specifically at images with the lungs and pathological features of COVID-19 removed to quantify the contribution of non-pathological features to model performance. This approach allows for a deeper analysis of model reliability without the availability of costly annotations.
Our main contributions of this paper are as follows:
-
An experimental review of top-cited COVID-19 models with carefully curated datasets to quantify and thoroughly evaluate the state of generalisability of COVID-19 CXR models.
-
Highlight the impact of pre-training strategies on generalisable learning, through the comparison of generic model weights against closely related weights. From this emerges further support for the use of network pre-training on closely related tasks rather than ImageNet.
-
We present a chest occlusion evaluation to quantify and approximate model reliance on non-pathological features (shortcut learning) without the need for costly annotations. Our results strongly suggest that models trained without closely related pre-training had higher dependence on shortcut features, which strongly correlates with poor generalisation capability. From this, we can infer that shortcut learning is a critical barrier to achieving robust model predictive performance. We recommend the incorporation of similar tests in model evaluations when external data is unavailable to assist in the identification of shortcut learning.
The remainder of this paper is structured as follows. Section 2 describes the specific challenges and criticisms of COVID-19 studies, Section 3 details our experiment and Section 4 describes our results. In Section 5 we summarise our findings and discuss the outcomes of this study.
2 Related work
Throughout the COVID-19 pandemic, the novel nature of the disease and strict regulations associated with sensitive medical data limited the amount of public data available for model training and evaluation, with many publications relying on the aggregate COVID-19 Image Data Collection [15]. Deep neural networks trained exclusively on small medical image datasets have been found to generalise poorly onto new datasets [22]. To overcome this limitation, transfer learning has been shown to help models achieve state-of-the-art performance when trained on limited datasets in mainstream image classification tasks. In their systematic review Roberts et al. [6], observed that many COVID-19 papers implemented transfer learning, using weights learned from the natural image dataset, ImageNet [3]. However, in a pre-COVID-19 study focused on pneumonia classification, Zech et al. [23] showed that models pre-trained on ImageNet did not consistently generalise onto external tests, despite achieving strong internal performance. Some papers performed model pre-training on general chest x-ray datasets, before fine-tuning on COVID-19 data [24, 25], however neither performed external evaluation tests, so the impact of transferring weights from a closely related task is unclear.
Critical reviews of the literature regarding COVID-19 models frequently shared concerns regarding reported model performance. Systematic reviews produced by the medical community [5, 6] highlighted the high risk of bias in datasets and models which, combined with the ambiguous reporting of results, has led to a lack of trust in models produced. Wynants et al. [5] warned that the use of aggregated COVID-19 datasets may lead to unintentional data overlap across training and test sets. This is evident in the literature, where several COVID-19 papers indicate the use of datasets with accidental data duplication [26,27,28]. Further concerns of data usage were raised in [6], where it was revealed that of the 37 papers evaluated, 29 performed no external validation of their models, without which, the reliability of predictive ability is difficult to determine. Similar concerns were raised in [29] and [14], where both reported on a lack of generalisation reporting in COVID-19 model literature. Roberts et al. [6] also highlighted the significant risk of participant bias, after identifying 16 publications which used paediatric CXRs as control images for COVID-19 classification [6]. The use of such datasets may lead to models becoming reliant on image features unrelated to pathology [8], thus making them inappropriate for clinical use.
To better understand the extent to which COVID-19 models may learn spurious correlations, a broader review of contextual works is required. The use of segmented images has been shown to be beneficial for models. In [14] it was demonstrated that using UNET to extract only the lungs from CXRs in conjunction with data augmentations limited the possibility for learning known CXR shortcuts. However, it has been observed that CXRs with extensive diseased regions can cause lung segmentation models to fail [30], leading to subsequent classification failure. Similarly, machine artifacts can confound segmentation models, and specialised methods are required to address these challenges [31]. As such, care must be taken when relying on image masking to alleviate shortcuts.
Regarding COVID-19 specifically, [32] demonstrated how training on aggregated datasets with class-wise stratification of sources, a common practice in COVID-19 literature, can facilitate the learning of irrelevant features. They also found that training on a single data source, while achieving a lower internal score than the aggregate models, produced a significantly stronger generalising model. Similar findings were found by [17], who trained models on two different composite datasets, each with class-wise stratification of data sources. In their cross-dataset evaluation, both models gave outstanding performance on internal tests but suffered severe performance drops when tested externally. Through a careful review of model saliency maps, they demonstrated how models were reliant on non-pathological features, such as laterality markers and shoulder positioning, and argued that this shortcut learning contributes to the generalisation failure of models. In a simple yet effective experiment, [33] showed that even with lung tissue occluded, some deep learning models can easily distinguish between data sources. Given the prevalence of COVID-19 models trained on datasets with class-wise stratification of sources, this sets a concerning precedent for the reliability of reported model performances.
The limited availability of medical imagery, and the intrinsic lack of explainability associated with deep learning models, has led to the use of saliency maps to validate model ability. The authors of [6] argue that the inclusion of such interpretative techniques is imperative for the clinical adoption of these technologies. However, in [34] there is a warning that the findings of such techniques can be misleading, as there is no guarantee that features identified in a highlighted region of interest correlate directly to the pathological finding. These concerns are supported by the work of [35], where different methods of mitigating model dependence on non-pathological features were evaluated. It was found that models presenting good saliency features did not always generalise well. Conversely, models with stronger generalisability did not always obtain good saliency attributions. As such, while saliency can be useful in revealing model reliance on inappropriate features, it should not be assumed that apparent good feature attributions will produce a strongly generalisable model, and they must not used in place of external validation.
Schematic diagram of our experiment. Images are first normalised to reduce noise and source biases. We then train a suite of model architectures with various pre-training strategies. Models are then evaluated on Internal and External datasets (shown in Fig. 3) to determine their generalisation ability. We then perform a chest occlusion test to understand how models behave when pathology-relevant regions are hidden, and determine whether this relates to their cross-dataset performance. The results are discussed in Section 4
3 Materials & methods
To determine the generalisability of deep learning models trained on a limited dataset, we performed an extensive cross-dataset evaluation. By using multiple external datasets, we can more reliably assess the ability of models to generalise beyond the training dataset. Furthermore, we can better understand the extent to which learned features are relevant across different datasets, which is essential for deploying such models in real-world applications. We also evaluate the impact of concealing a known region of interest, the lung tissue, to determine model reliance on irrelevant features. Models which continue to distinguish between classes, despite the true pathology being hidden, are likely to generalise poorly. We compare the results of this test against the cross-dataset stability of the models. A visual summary of our study is shown in Fig. 2. The following subsections detail our data curation efforts, describing the characteristics of each data source and the justification for our inclusion criteria.
3.1 Datasets
In this study, we consider four distinct datasets, one for training and internal testing. The protocol for gathering these datasets has been carefully considered, taking into account the precedence of the source datasets to ensure as little as possible overlap between different datasets. The composition of each is shown in Fig. 3.
3.1.1 Internal
The covid-xray-5k dataset [18] was used for model training and testing. We refer to this as the Internal dataset. It combines COVID-19 images from the covid-chestxray-dataset [15] and non-COVID samples from CheXpert [36]. covid-chestxray was one of the first publicly available COVID-19 datasets and is one of the most commonly used in COVID-19 model literature. It contains images collected from medical websites and publications, as well as user-submitted images, resulting in a broad range of image variations. Image formats include both JPEG and PNG. In contrast, the non-COVID CheXpert images were obtained exclusively from Stanford Hospital in California, USA, between 2002 and 2017. All CXRs were filtered to only contain Anterior-Posterior (AP) or Posterior-Anterior (PA) view images. We used the predefined train and test split for this dataset, which has a 33-67% split. The classes are heavily imbalanced as highlighted in Fig. 4, with a total 184 COVID-19 images and 5000 negative images. All images are shared as JPEGs.
3.1.2 External I
Since the publication of covid-xray-5k [18], further 152 AP/PA COVID-19 images have been added to the covid-chestxray-dataset [15]. We combined these newly available images with normal and pneumonia CXRs from the BRAX dataset [37] to produce our first test set, External dataset I. The BRAX dataset consists of CXRs collected from the the Hospital Israelita Albert Einstein, Brazil between 2008 and 2017. Images were automatically extracted from radiology reports using the CheXpert [12] label extraction algorithm. The original dataset contains over 40,000 images classified across 14 different findings labels. The authors shared both DICOM and PNG versions of the images, however we considered only the PNGs for this study. We randomly sampled 152 Normal and 152 Pneumonia images from the dataset, ensuring images were all either PA or AP view.
To prevent potential overlap and duplication of COVID-19 samples between External I and the Internal datasets, images present in the Internal datasets were carefully removed using filename filtering. Additionally, we compared the URL sources of images of both datasets to check for overlap. We observed overlap of sources from radiopaedia.org, sirm.org, eurorad.org, rsna.org and sciencedirect.com. All of these sites are composite sources; images are either collected from journal publications or user-submitted, hailing from countless institutions. As such, these overlaps are less likely to be as severe as initially observed. Furthermore, 17 new data sources are introduced into the External I dataset, thus ensuring new sites are being evaluated. Despite this, due to the eclectic nature of this data collection, detailed image meta-data is not always available. The ambiguity of image origins, such as hospital and patient details, means this dataset cannot be guaranteed to be completely mutually exclusive of the Internal dataset, therefore we consider this dataset as a weak external validation. To address the limitation of this dataset, we created an additional two datasets for more realistic external model evaluations.
3.1.3 External II
For the second external evaluation dataset, we used the SIIM-FIABIO RSNA COVID-19 dataset [38]. This combines images from the BIMCV COVID-19+ dataset [39] with additional COVID-19 samples from the RSNA RICORD dataset [40]. Unlike the previous external dataset, all images in this dataset were sourced from controlled, clinical environments. BIMCV images were obtained from the Valencian Region Medical Image Bank, which includes data from 11 hospitals throughout the Valencian region of Spain. The RSNA RICORD dataset contains COVID-19 images from medical institutions in Turkey, the USA, Canada and Brazil. Both datasets provided images as high-resolution DICOM files. The preprocessing steps applied to the External I DICOM images, were repeated for these images. Using the predefined test set, External II contains 1752 COVID-19 images and 671 non-COVID-19 images. In contrast to the previous datasets, this dataset contains a vast amount of COVID-19 images and significantly fewer negative images.
3.1.4 External III
External III is our final external evaluation set and is comprised of images exclusively from the COVID-GR dataset [41]. It is one of the only COVID-19 datasets that provides an even distribution of both COVID-19 positive and negative CXRs collected within the same timeframe from a single institution. Images were procured from the Hospital Universitario Clinico San Cecilio, Granada, Spain. It contains 426 positive and 426 negative COVID-19 CXRs, which are exclusively PA views.
External datasets II and III were shared by medical institutions. As these images come from a controlled and consistent source, they are likely to better represent data in applied clinical scenarios, compared to the mass aggregated COVID-19 samples used in Internal and External I. As such, the discriminative ability of models on these datasets is of particular interest.
3.1.5 Processing
As can be seen in Fig. 5, image brightness and contrast can vary greatly between images. These variations may occur naturally in a dataset, such as the use of different machines and the quality of the x-ray. These variations, which are irrelevant to the classification target, introduce noise into the dataset, making it more challenging for a model to learn the important features of an image. To create more consistency across the images, we applied histogram equalisation to all images using the OpenCV image processing library for Python. This process aims to redistribute the grey levels throughout the image more evenly and improve image contrast. Crucially, unlike adaptive histogram equalisation techniques, the original shape of the distribution is kept. This maintains tissue density information, which can be determined by comparing the intensity of different tissues. We evaluated all models on the original and the histogram equalised images, then report the results from the best image type for that architecture. We specify which image types were used for each model in Section 3.2.
Examples of natural variations in chest x-rays from the BIMCV dataset [39]
3.1.6 Occlusion test
Influenced by the analysis produced by Maguolo et al. [33], we introduce a novel occlusion test (see Fig. 6) to help determine whether models which fail this internal test are more prone to generalisation failure. Using the test images from Internal, we produced a new occlusion test set. This was achieved by placing a black box directly over the dark lung tissue, thus concealing the crucial chest region (shown in Fig. 6). This process should conceal all pathology features which may be present, but does not rely on expert annotators. As it could be said that any drop in performance could simply be due to an effective drop in the number of pixels available for analysis, we also include a control dataset where a random region of each image is occluded with a black box. We then tested each model on the occluded dataset (chest and control) to determine whether they could still identify COVID-19. A model which could still distinguish easily between classes on images where the lung region is occluded is likely to depend on pathology-irrelevant features, and may be subject to shortcut learning. Conversely, a model which fails to distinguish between classes after lung occlusion may be more reliant on relevant features. We quantify this assumption by examining the difference between the number of accurate predictions made by models on both the lung-occluded dataset and the control-occluded dataset and comparing this against the standard deviation of the models when tested on all datasets.
3.1.7 Class distribution
Severe class imbalance, as found in the training data, can lead to unreliable, biased models. To prevent this behaviour, many COVID-19 publications implemented a variety of class-balancing techniques to encourage fair learning between classes. In this study, we apply class weighting to our models, but do not apply class weighting to reproduced models (DarkCovidNet and DeepCOVID). We chose this technique as it does not produce augmented data samples. Without review by a radiologist, techniques which modify images to produce new samples, such as data augmentation and generative adversarial networks (GANs), may result in invalid images where important pathological features are obscured or completely destroyed. Class weighting assigns weights to the model loss function during training, to encourage it to assign greater importance to the minority class samples. In our class weighted models, we assigned the negative class a weight of 0.5195 and the COVID-19 class 13.3205 using the following equation:
where \(n_{class}\) is the total number of samples of the class in the dataset and \(n_{data}\) is the total number of images in the dataset and the specified class weight is denoted as \(W_{class}\).
3.1.8 Evaluation
We compare the cross-dataset performance of the models using the area under the receiver operating characteristic curve (AUC). AUC ranges in values from 0 to 1, where 0.5 is considered random guessing and 1 indicates a perfect ability to distinguish between classes. In cross-dataset evaluations, we report the AUC of each dataset, as well as the model stability, \(SD_{all}\). We quantify model stability as the standard deviation of the model AUC across all evaluated datasets. Stable models are models which maintain their performance across multiple datasets and can therefore be said to generalise well, even if their performance on the internal dataset is not state of the art. As model stability increases, the standard deviation of AUC should lower. We also calculate the specificity and precision of models when tuned to a sensitivity value of 0.98 due to the importance of always identifying positive cases. However, we only report the specificity and precision on the internal test, as models with little to no discriminative ability in external tests produce insignificant values when tuned to high sensitivity thresholds. Confidence intervals (CI) at 0.98 of three model runs are provided on internal tests.
3.2 Models
We consider four deep learning architectures. These comprise: the novel COVID-19 architecture DarkCovidNet [11]; DenseNet-121 [42] with three sets of pre-trained weights; a reproduction of the Deep-COVID ResNet-16 proposed by [18]; and a four-convolutional layer network (Conv-4) to serve as a comparative baseline.
DenseNets are deep neural networks comprised of convolutional and average pooling layers. Each layer is connected to all the previous layers, creating a highly dense network. They have been reported as one of the best architectures for both general chest x-ray classification [43] and COVID-19 [6]. Due to the popularity of DenseNets, we were able to obtain not only architecture weights from the popular ImageNet [3] benchmarking dataset, but also novel COVID-19 weights shared by [24]. We used three variations of DenseNet.
The first variation of DenseNet was initialised with ImageNet weights. While the ImageNet dataset does not contain images related to medical imagery, it is well established that fine-tuning ImageNet model weights on new tasks can produce state-of-the-art performance, even when fine-tuning with relatively few new images [44]. As such, this approach has been used extensively in the COVID-19 literature.
Our second variation of DenseNet was based upon the work shared by Wehbe et al. [24]. Few COVID-19 models were pre-trained on medical images or greyscale datasets. [24] was one of the few, who used a general chest x-ray dataset, CXR-8 [12], to pre-train their models. This prior exposure to a single-source CXR dataset may facilitate the learning of general chest features. The Wehbe models were originally fine-tuned on a vast private clinical COVID-19 dataset containing over 3000 COVID-19 samples. While this dataset is not publicly available, by using the shared model weights, we can take advantage of the additional data. Incorporating model weights which contain COVID-19 information may encourage the learning of generalisable features, thus producing a better generalising model. To determine the impact of pre-training on a closely related dataset versus simple exposure to more data, we trained our own third variation of DenseNet with the same CXR-8 dataset [12] as used on the Wehbe model [24]. Using the pre-defined dataset train and test split, we first initialised a DenseNet model with ImageNet weights, then fine-tuned the network on the CXR-8 dataset. In our initial attempts, DenseNet struggled to converge on the full, multi-class dataset. Therefore, we simplified the task to binary classification between pneumonia and normal classes. There was a severe class imbalance present, with more than 60,000 normal images against a mere 1431 pneumonia. We randomly under-sampled the normal class to 3000 samples and applied histogram equalisation to all images. When fine-tuning, we first trained only the output layer for 50 epochs, then unfroze all network layers and continued training for 10 epochs. The model weights were then saved.
All DenseNet-121 models were subsequently fine-tuned on Internal I in a consistent manner. First, the transfer weights were loaded into the network, and the final layer was replaced with a single-unit Dense layer. Initial training was achieved by freezing all model layers, except the output, and trained for 50 epochs with a learning rate of 0.001, to tune the classification layer. The model was then fine-tuned by unfreezing all layers and training for a further 10 epochs and a lower learning rate of 0.0001. The best weights were then saved and used for evaluation.
Following on from our DenseNet variations, the second architecture we consider is DarkCovidNet, proposed by [11]. The authors modified the state-of-the-art DarkNet architecture [45], to focus on small differences in images, rather than the image as a whole. The authors argue this modification can allow the model to better recognise subtle pathological features than typical networks. We used the code shared by the authors to train a DarkCovidNet on our internal dataset. Unlike the previous models, whose input dimensions were set to the typical 224x224, DarkCovidNet uses image dimensions of 256x256. We compared the performance of the model using the original images and histogram equalised images, and found the original images produced slightly better performance. As such, we share the results of the model trained and tested on the original images. Additionally, given that all other models in this study were trained on oversampled data or with class weights, we also compared the performance of this architecture with oversampling. We observed no significant improvement in model performance, and therefore used the original model, as shared by the authors.
In their Deep-COVID paper, [18] experimented with various architectures, however in this study we only evaluate the model for which they shared code: ResNet-18 [46]. We reproduced their model, transferring weights from ImageNet and then fine-tuned the final output layer of the network on COVID-19 data (Internal dataset). We compared the performance of the model trained for 100 epochs, as performed in their study, and 50 epochs, finding no significant difference in performance. Therefore, we report the results of the model trained over 50 epochs for consistency with other models. As performed on DarkCovidNet, we compared the performance of the model when trained using histogram equalised and original images, and observed stronger performance using the original images. As such, we report the results of the original images.
Finally, we introduce a simple deep neural network architecture, similar to LeNet-5 [47] and AlexNet [48], to serve as a simple baseline model for comparison. It consists of four convolutional layers with a kernel size of (3, 3), each followed by a max-pooling layer. These are flattened and followed by two fully connected layers, the final layer outputting the probability of the image being positive for COVID-19. We used LeakyReLU layer activations with an alpha value of 0.2. Models were trained for a maximum of 50 epochs with a batch size of 32.
Models were produced using the Keras framework [49]. An early stopping policy was included in the Keras models that ended model training if there was no improvement in training loss for 30 epochs. The policy also restored the best-performing weights from all epochs. We used Stochastic Gradient Descent (SGD) and cross-entropy loss for model optimisation.
4 Results
In Table 1, we present the internal performance metrics of the six models. DarkCovidNet achieves outstanding results, demonstrating a perfect ability to distinguish between classes across all metrics. Most models report excellent AUC scores however, the CXR pre-trained networks, namely CXR-8 and Wehbe, score significantly lower.
Alarmingly, the CXR-8 model scores the lowest of all, achieving an AUC of 0.629. When tuned to 98% sensitivity, only DarkCovidNet achieves high specificity and precision scores. The poor precision scores indicate that the models are biased towards the COVID-19 class. Reviewing these metrics alone, one would conclude that DarkCovidNet is the most reliable model, and that the CXR-8 pre-trained network is the worst. However, the results of the cross-dataset evaluation reveal a different story.
Turning to the results of our cross-dataset evaluation, as shown in Table 2, it becomes clear that the internal scores are not accurately representative of the applied performance of the models. Highlighted in Fig. 7, the best performing models from the Internal test tend to suffer the worst performance degradation when tested externally. While generally scoring lower in internal tests, the CXR pre-trained networks consistently achieve the strongest AUCs on external datasets, maintaining small standard deviations (SD) of less than 0.1 across all datasets.
The only exception is on External I, where the ImageNet and Wehbe models report the highest AUC scores, followed by CXR-8 with a relatively poor AUC of 0.672. While a low score, it still outperforms the other architectures, and it achieves the second strongest AUCs on both external II and III, surpassing its internal performance.
Most surprising is the dramatic generalisation failure of DarkCovidNet, which despite achieving exemplary performance on the internal test, displays little to no ability to distinguish between classes, with a large AUC SD of 0.232. The external scores are comparable to Conv-4, our baseline model. Deep-COVID performs slightly better in external tests, indicating some discriminative ability, however, these scores are still poor and markedly lower than its internal performance. Overall, these results show that pre-training on a closely related task can significantly improve the performance of models on external datasets, even when fine-tuning on a challenging dataset.
Bar plot visualising model performance in occlusion evaluation. Statistically significant predictive ability of 0.5 AUC is denoted by the red dashed line. Black bars on the Chest occluded results visualise performance drop from Control to Chest occluded images. Orange bars indicate when performance has dropped to below 0.5
These findings are supported by the results of our occlusion test, as shown in Table 3. Random occlusion causes a small drop in performance across all models. In contrast, chest occlusion has a much more varied impact on the different models. Figure 8 clearly illustrates that CXR pre-trained models experience a significant decrease in performance when the lung tissue is hidden, with AUCs dropping to values within 0.15 of random guessing (0.5).
The other models generally maintain AUCs around 0.8 and higher, indicating their ability to distinguish between classes, despite true pathology features being hidden. Interestingly, ImageNet pre-training reports one of the smallest drops in performance from Control to Chest occlusion (0.04 AUC), indicating these models rely almost exclusively on features outwith the lung tissue.
Reliance on features from non-pathology regions appears to align with model stability, as indicated by the SD of AUC. Models with lower stability performed more poorly on external datasets, indicating a lack of generalisation capacity. These results suggest that transferring closely related weights not only improves model generalisation, but may also encourage models to focus on learning relevant features closer to the pathology features that human radiologists utilise. This, in turn, supports the generally held belief that while deep learning models can perform better on highly specific tasks, humans remain better at generalisation.
Finally, the performance of the pre-trained Wehbe model surpasses all others in our study however, as it has been exposed to another controlled COVID-19 dataset, any performance benefits using these weights could be attributed to this additional data. To determine the impact of the original weights, we compare the original Wehbe model weights as shared by the authors, against both the CXR-8 model weights and the Wehbe model weights fine-tuned on the internal dataset. The results are shown in Table 4. Surprisingly, the original Wehbe performs significantly poorer than our fine-tuned version and only outperforms the CXR-8 model on the internal test set. However, despite the weaker performance, the original Wehbe weights exhibit similar cross-dataset stability to the other CXR pre-trained networks. Combined with the previous results, we can infer that network pre-training on a closely related task can help prevent a model from aggressively turning to easy but irrelevant distinguishing features in the data. However, as shown by the original Wehbe model weights, this does not guarantee the model will perform well overall. Exposing these models to more diverse data, as with our fine-tuned Wehbe model, appears to significantly improve model generalisability, even when the dataset has proven challenging.
In summary, the results lead us to the vital recommendation that computer vision models intended for medical applications adopt a robust and reproducible approach to assessing generalisation. These experiments show that performance gains attributed to novel model architectures may actually be enhancing shortcut learning and thus undermining any potential gains in a general setting.
5 Conclusions
In this paper, we performed a generalisation study on chest x-ray classifiers for the detection of COVID-19. Since the COVID-19 publication boom, additional clinical datasets have become publicly available. As such, we were able to thoroughly analyse the applied performance and generalisation capacity of high-impact and heavily cited methodologies. Specifically, we evaluated the performance of models when trained on a challenging dataset that faithfully represents those available during the pandemic, but were tested on a greater variety of datasets. This allowed us to establish whether specific techniques could improve CXR model generalisation. We also introduced a chest occlusion evaluation to determine model reliance on known shortcut features without radiologist annotations.
The results of this study reveal a concerning pattern; that models can achieve state-of-the-art performance in internal tests but experience severe performance degradation in external evaluations. Some highly-cited models displayed symptoms of shortcut learning, with discriminative ability on external datasets close to random guessing. We also observed that while models trained on a closely related task did not always achieve state-of-the-art performance, they proved to be more stable, performing almost as well on new datasets as they did on their training distribution. Furthermore, architecture choice apparently had little impact on performance, with bespoke COVID-19 architectures generalising as poorly as a relatively simple CNN. One of the more significant findings to emerge is the marked improvement of shortcut robustness when implementing specialised CXR pre-training strategies. Furthermore, the chest occlusion test can share useful insights to model ability when external evaluation data is unavailable. Our results suggest that models with strong predictive ability in the occlusion test are likely to produce poorly generalising models. However, the generalisability of these findings are subject to certain limitations. For instance, due to the limited internal dataset size, manual generation of occlusion images was feasible. On larger test sets, an automated process would likely be required. The use of lung segmentation models may produce inaccurate occlusion boxes on heavily diseased CXRs, leading to inaccurate evaluations. Similarly, due to the shape of the occlusion box, it may also occlude known shortcuts, such as laterality markers. Finally, our occlusion test can only report on spurious feature reliance outside the lung tissue region. Non-pathology indicators may still be present in the lung tissue, and this evaluation can only approximate reliance outside the lung tissue without additional radiological annotations.
We have established concerning issues regarding how deep learning models have been trained for the detection of COVID-19, and the consequent severe generalisation failure. Going forward, we must consider how best to approach these challenges beyond COVID-19 classification to medical image models as a whole. The incorporation of cross-dataset evaluation into model publications is paramount to address the significant risk of unreliable, dataset-specific biases and to determine the applied performance of models. In this paper, we have supplied a protocol for the use of existing datasets such that generalisation testing is reproducible. Although generalisation testing will likely slow down the development of exceptional accuracy, it is vital to encourage the adoption of the latest deep learning techniques into the medical arena.
The results of this study indicate that a combination of controlled, clinical and diverse datasets are necessary for improving model generalisation. However, relatively small samples of each from closely related tasks can contribute greatly to model stability. Recent works have proposed novel methods of guiding networks to regions of interest, which may help mitigate the extent to which models rely on spurious correlations. For example, [50] used lung segmentation networks to square crop CXRs to the minimal required region, which helped boost their architecture performance in their internal evaluations. However, the authors observed severe segmentation failure in severe cases with many lung opacities, which led to classification failure. Alternatively, [51] incorporated radiologist eye-tracking into their model, to focus on known regions of interest. Future work must incorporate validation of these models on external datasets, so that the impact of such features on model generalisation can be quantified. Achieving reliable learning is foundational for success in this domain, but progress here will positively impact broader applications.
Further research is required to determine the efficacy of focused network pre-training on more challenging classification tasks, such as broader chest x-ray classification, as well as different forms of imagery. Additionally, methods which artificially introduce diversity to datasets, such as GANs and data augmentation, should be investigated to determine whether they can effectively enrich limited datasets and improve generalisation when used for fine-tuning.
Data Availability
The datasets and models used in this study are available in the accompanying Generalisable COVID-19 repository, https://github.com/sophie-haynes/Gen_COVID_Challenges.
References
Kim HE, Cosa-Linan A, Santhanam N, Jannesari M, Maros ME, Ganslandt T (2022) Transfer learning for medical image classification: A literature review. BMC Med Imaging 22(1):69. https://doi.org/10.1109/CVPR.2011.5995347
Torralba A, Efros AA (2011) Unbiased look at dataset bias. In: CVPR 2011, IEEE, pp 1521–1528
Deng J, Dong W, Socher R, Li L-J, Li K, Fei-Fei L (2009) Imagenet: A large-scale hierarchical image database. In: 2009 IEEE Conference on computer vision and pattern recognition. https://doi.org/10.1109/CVPR.2009.5206848
Li Z, Evtimov I, Gordo A, Hazirbas C, Hassner T, Ferrer CC, Xu C, Ibrahim M (2023) A Whac-A-Mole Dilemma: Shortcuts Come in Multiples Where Mitigating One Amplifies Others. CVF/IEEE Conf Comput Vis Pattern Recogn (CVPR). arXiv:2212.04825
Wynants L, Van Calster B, Collins GS, Riley RD, Heinze G, Schuit E, Bonten MMJ, Dahly DL, Damen JA, Debray TPA, Jong VMT, De Vos M, Dhiman P, Haller MC, Harhay MO, Henckaerts L, Heus P, Kammer M, Kreuzberger N, Lohmann A, Luijken K, Ma J, Martin GP, McLernon DJ, Andaur Navarro CL, Reitsma JB, Sergeant JC, Shi C, Skoetz N, Smits LJM, Snell KIE, Sperrin M, Spijker R, Steyerberg EW, Takada T, Tzoulaki I, Kuijk SMJ, Bussel BCT, Horst ICC, Royen FS, Verbakel JY, Wallisch C, Wilkinson J, Wolff R, Hooft L, Moons KGM, Smeden M (2020) Prediction models for diagnosis and prognosis of covid-19: systematic review and critical appraisal, BMJ. https://doi.org/10.1136/bmj.m1328
Roberts M, Driggs D, Thorpe M, Gilbey J, Yeung M, Ursprung S, Aviles-Rivero AI, Etmann C, McCague C, Beer L, Weir-McCall JR, Teng Z, Gkrania-Klotsas E, Ruggiero A, Korhonen A, Jefferson E, Ako E, Langs G, Gozaliasl G, Yang G, Prosch H, Preller J, Stanczuk J, Tang J, Hofmanninger J, Babar J, Sánchez LE, Thillai M, Gonzalez PM, Teare P, Zhu X, Patel M, Cafolla C, Azadbakht H, Jacob J, Lowe J, Zhang K, Bradley K, Wassin M, Holzer M, Ji K, Ortet MD, Ai T, Walton N, Lio P, Stranks S, Shadbahr T, Lin W, Zha Y, Niu Z, Rudd JHF, Sala E, Schönlieb CB (2021) Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nature Mach Intell (3). https://doi.org/10.1038/s42256-021-00307-0
Wong HYF, Lam HYS, Fong AH-T, Leung ST, Chin TW-Y, Lo CSY, Lui MM-S, Lee JCY, Chiu KW-H, Chung TW-H, Lee EYP, Wan EYF, Hung IFN, Lam TPW, Kuo MD, Ng M-Y (2020) Frequency and Distribution of Chest Radiographic Findings in Patients Positive for COVID-19. Radiology. https://doi.org/10.1148/radiol.2020201160
Tartaglione E, Barbano CA, Berzovini C, Calandri M, Grangetto M (2020) Unveiling COVID-19 from CHEST X-Ray with Deep Learning: A Hurdles Race with Small Data. Int J Environ Res Public Health (18). https://doi.org/10.3390/IJERPH17186933
Ng MY, Lee EYP, Yang J, Yang F, Li X, Wang H, Lui MMS, Lo CSY, Leung B, Khong PL, Hui CKM, Yuen KY, Kuo MD (2020) Imaging Profile of the COVID-19 Infection: Radiologic Findings and Literature Review. Radiol Cardiothorac Imaging (1). https://doi.org/10.1148/RYCT.2020200034
Castiglioni I, Ippolito D, Interlenghi M, Monti CB, Salvatore C, Schiaffino S, Polidori A, Gandola D, Messa C, Sardanelli F, Castiglioni I (2020) Artificial intelligence applied on chest X-ray can aid in the diagnosis of COVID-19 infection: a first experience from Lombardy. Italy medRxiv. https://doi.org/10.1101/2020.04.08.20040907
Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Rajendra Acharya U (2020) Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. https://doi.org/10.1016/j.compbiomed.2020.103792
Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM (2017) ChestX-ray8: Hospital-scale Chest X-ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases. In: Proceedings of the IEEE conference on computer vision and pattern recognition (CVPR). https://doi.org/10.1109/CVPR.2017.369
Wang L, Lin ZQ, Wong A (2020) COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci Rep (1). https://doi.org/10.1038/s41598-020-76550-z
Ahmed KB, Goldgof GM, Paul R, Goldgof DB, Hall LO (2021) Discovery of a generalization gap of convolutional neural networks on covid-19 x-rays classification. IEEE Access 9:72970–72979. https://doi.org/10.1109/access.2021.3079716
Cohen JP, Morrison P, Dao L, Roth K, Duong TQ, Ghassemi M (2020) COVID-19 Image Data Collection: Prospective Predictions Are the Future. Mach Learn Biomed Imaging. https://doi.org/10.48550/arXiv.2006.11988
Kermany DS, Goldbaum M, Cai W, Valentim CCS, Liang H, Baxter SL, McKeown A, Yang G, Wu X, Yan F, Dong J, Prasadha MK, Pei J, Ting M, Zhu J, Li C, Hewett S, Dong J, Ziyar I, Shi A, Zhang R, Zheng L, Hou R, Shi W, Fu X, Duan Y, Huu VAN, Wen C, Zhang ED, Zhang CL, Li O, Wang X, Singer MA, Sun X, Xu J, Tafreshi A, Lewis MA, Xia H, Zhang K (2018) Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell (5). https://doi.org/10.1016/J.CELL.2018.02.010
DeGrave AJ, Janizek JD, Lee SI (2021) AI for radiographic COVID-19 detection selects shortcuts over signal. Nature Mach Intell 2021 3:7 (7). https://doi.org/10.1038/s42256-021-00338-7
Minaee S, Kafieh R, Sonka M, Yazdani S, Jamalipour Soufi G (2020) Deep-COVID: Predicting COVID-19 from chest X-ray images using deep transfer learning. Med Image Anal. https://doi.org/10.1016/j.media.2020.101794
Geirhos R, Jacobsen J-H, Michaelis C, Zemel R, Brendel W, Bethge M, Wichmann FA (2020) Shortcut learning in deep neural networks. Nature Machine Intelligence 2(11):665–673. https://doi.org/10.1038/s42256-020-00257-z
Ahmed KB, Hall LO, Goldgof DB, Fogarty R (2022) Achieving multisite generalization for cnn-based disease diagnosis models by mitigating shortcut learning. IEEE Access 10:78726–78738. https://doi.org/10.1109/ACCESS.2022.3193700
de Sousa Freire N, de Souza Leo PP, Tiago LA, de Almeida Campos Gonalves A, Pinto RA, dos Santos EM, Souto E (2023) Analysis of generalizability on predicting COVID-19 from chest X-ray images using pre-trained deep models. Comput Methods Biomech Biomed Eng Imaging Vis 0(0):1–11. https://doi.org/10.1080/21681163.2023.2264408
Abbas A, Abdelsamea MM, Gaber MM (2021) Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl Intell (2). https://doi.org/10.1007/S10489-020-01829-7
Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, Oermann EK (2018) Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: A cross-sectional study. PLOS Medicine (11). https://doi.org/10.1371/journal.pmed.1002683
Wehbe RM, Sheng J, Dutta S, Chai S, Dravid A, Barutcu S, Wu Y, Cantrell DR, Xiao N, Allen BD, MacNealy GA, Savas H, Agrawal R, Parekh N, Katsaggelos AK (2021) DeepCOVID-XR: An artificial intelligence algorithm to detect COVID-19 on chest radiographs trained and tested on a large U.S. Clinical data set. Radiology (1). https://doi.org/10.1148/radiol.2020203511
Cohen JP, Dao L, Morrison P, Roth K, Bengio Y, Shen B, Abbasi A, Hoshmand-Kochi M, Ghassemi M, Li H, Duong TQ (2020) Predicting COVID-19 Pneumonia Severity on Chest X-ray with Deep Learning. Int J Comput Appl (7). https://doi.org/10.5120/ijca2021921353
Khan AI, Shah JL, Bhat MM (2020) CoroNet: A deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput Methods Programs Biomed. https://doi.org/10.1016/j.cmpb.2020.105581
Hussain E, Hasan M, Rahman MA, Lee I, Tamanna T, Parvez MZ (2021) CoroDet: A deep learning based classification for COVID-19 detection using chest X-ray images. Chaos, Solitons & Fractals. https://doi.org/10.1016/j.chaos.2020.110495
Ismael AM, Sengür A (2021) Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Syst Appl. https://doi.org/10.1016/j.eswa.2020.114054
López-Cabrera JD, Orozco-Morales R, Portal-Diaz JA, Lovelle-Enríquez O, Pérez-Díaz M (2021) Current limitations to identify COVID-19 using artificial intelligence with chest X-ray imaging. Health and Technology (2). https://doi.org/10.1007/s12553-021-00520-2
Oh Y, Park S, Ye JC (2020) Deep Learning COVID-19 Features on CXR Using Limited Training Data Sets. IEEE Trans Med Imaging 39(8):2688–2700. https://doi.org/10.1109/TMI.2020.2993291
Shu X, Yang Y, Liu J, Chang X, Wu B (2023) Alvls: Adaptive local variances-based levelset framework for medical images segmentation. Pattern Recogn 136:109257. https://doi.org/10.1016/j.patcog.2022.109257
Dhont J, Wolfs C, Verhaegen F (2022) Automatic coronavirus disease 2019 diagnosis based on chest radiography and deep learning - Success story or dataset bias? Medical Physics (2). https://doi.org/10.1002/MP.15419
Maguolo G, Nanni L (2021) A critic evaluation of methods for COVID-19 automatic detection from X-ray images. Information Fusion. https://doi.org/10.1016/j.inffus.2021.04.008
Ghassemi M, Oakden-Rayner L, Beam AL (2021) The false hope of current approaches to explainable artificial intelligence in health care. The Lancet Digital Health (11). https://doi.org/10.1016/S2589-7500(21)00208-9
Viviano JD, Simpson B, Dutil F, Bengio Y, Paul Cohen J (2021) Saliency is a Possible Red Herring When Diagnosing Poor Generalization. In: International conference on learning representations. https://doi.org/10.48550/arXiv.1910.00199
Irvin J, Rajpurkar P, Ko M, Yu Y, Ciurea-Ilcus S, Chute C, Marklund H, Haghgoo B, Ball R, Shpanskaya K, Seekins J, Mong DA, Halabi SS, Sandberg JK, Jones R, Larson DB, Langlotz CP, Patel BN, Lungren MP, Ng AY (2019) CheXpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert Comparison. AAAI https://doi.org/10.48550/arXiv.1901.07031
Reis EP, Paiva J, Carolina M, Silva B, Ribeiro GAS, Paiva VF, Bulgarelli L, Lee H, Santos PV, Brito V, Amaral L, Beraldo G, Filho JNH, Teles G, Szarf G, Pollard T, Johnson A, Celi LA, Amaro E (2022) BRAX, a Brazilian labeled chest X-ray dataset (version 1.0.0). PhysioNet. https://doi.org/10.13026/ae9a-f727
Lakhani P, Mongan J, Singhal C, Zhou Q, Andriole KP, Auffermann WF, Prasanna P, Pham T, Peterson M, Bergquist PJ al (2021) The 2021 siim-fisabio-rsna machine learning covid-19 challenge: Annotation and standard exam classification of covid-19 chest radiographs. https://doi.org/10.31219/osf.io/532ek
Iglesia Vayá M, Saborit JM, Montell JA, Pertusa A, Bustos A, Cazorla M, Galant J, Barber X, Orozco-Beltrán D, García-García F, Caparrós M, González G, Salinas JM (2020) BIMCV COVID-19+: a large annotated dataset of RX and CT images from COVID-19 patients. https://doi.org/10.48550/ARXIV.2006.01174
Tsai EB, Simpson S, Lungren MP, Hershman M, Roshkovan L, Colak E, Erickson BJ, Shih G, Stein A, Kalpathy-Cramer J, Shen J, Hafez M, John S, Rajiah P, Pogatchnik BP, Mongan J, Altinmakas E, Ranschaert ER, Kitamura FC, Topff L, Moy L, Kanne JP, Wu CC (2021) The rsna international covid-19 open radiology database (ricord). Radiology. https://doi.org/10.1148/radiol.2021203957
Tabik S, Gómez-Ríos A, Martín-Rodríguez JL, Sevillano-García I, Rey-Area M, Charte D, Guirado E, Suárez JL, Luengo J, Valero-González MA, García-Villanova P, Olmedo-Sánchez E, Herrera F (2020) Covidgr dataset and covid-sdnet methodology for predicting covid-19 based on chest x-ray images. IEEE J Biomed Health Inform (2020) https://doi.org/10.1109/JBHI.2020.3037127
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700–4708
Cohen JP, Hashir M, Brooks R, Bertrand H (2020) On the limits of cross-domain generalization in automated x-ray prediction. In: Medical imaging with deep learning, PMLR, pp 136–155. https://doi.org/10.48550/arXiv.2002.02497
He K, Girshick R, Dollár P (2019) Rethinking ImageNet Pre-Training. In: 2019 IEEE/CVF International conference on computer vision (ICCV). https://doi.org/10.1109/ICCV.2019.00502
Redmon J, Farhadi A (2017) Yolo9000: Better, faster, stronger. In: Proceedings of the IEEE conference on computer vision and pattern recognition (CVPR). https://doi.org/10.1109/CVPR.2017.690
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition. https://doi.org/10.1109/CVPR.2016.90
LeCun Y, Bottou L, Bengio Y, Haffner P (1998) Gradient-based learning applied to document recognition. Proceedings of the IEEE (11). https://doi.org/10.1109/5.726791
Krizhevsky A, Sutskever I, Hinton GE (2017) Imagenet classification with deep convolutional neural networks. Commun ACM (6) https://doi.org/10.1145/3065386
Chollet F (2015) Keras. GitHub. https://github.com/fchollet/keras
Cores D, Vila-Blanco N, Pérez-Alarcón M, Martínez-de-Alegría A, Mucientes M, Carreira MJ (2022) A few-shot approach for covid-19 screening in standard and portable chest x-ray images. Scie Rep 12:21511 (2022) https://doi.org/10.1038/s41598-022-25754-6
Sonsbeek T, Zhen X, Mahapatra D, Worring M (2023) Probabilistic integration of object level annotations in chest x-ray classification, pp 3630–3640. https://doi.org/10.48550/arXiv.2210.06980
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Haynes, S.C., Johnston, P. & Elyan, E. Generalisation challenges in deep learning models for medical imagery: insights from external validation of COVID-19 classifiers. Multimed Tools Appl 83, 76753–76772 (2024). https://doi.org/10.1007/s11042-024-18543-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-024-18543-y