Abstract
Liver disease in patients is on the rise due to environmental factors like toxic gas exposure, contaminated food, drug interactions, and excessive alcohol use. Therefore, diagnosing liver disease is crucial for saving lives and managing the condition effectively. In this paper, a new method called Liver Patients Detection Strategy (LPDS) is proposed for diagnosing liver disease in patients from laboratory data alone. The three main parts of LPDS are data preprocessing, feature selection, and detection. The data from the patient is processed, and any anomalies are removed during this stage. Then, during feature selection phase, the most helpful features are chosen. A novel method is proposed to choose the most relevant features during the feature selection stage. The formal name for this method is IB2OA, which stands for Improved Binary Butterfly Optimization Algorithm. There are two steps to IB2OA, which are; Primary Selection (PS) step and Final Selection (FS) step. This paper presents two enhancements. The first is Information Gain (IG) approach, which is used for initial feature reduction. The second is implementing BOA's initialization with Optimization Based on Opposition (OBO). Finally, five different classifiers, which are Support Vector Machine (SVM), K-Nearest Neighbor (KNN), Naive Bayes (NB), Decision Tree (DT), and Random Forest (RF) are used to identify patients with liver disease during the detection phase. Results from a battery of experiments show that the proposed IB2OA outperforms the state-of-the-art methods in terms of precision, accuracy, recall, and F-score. In addition, when compared to the state-of-the-art, the proposed model's average selected features score is 4.425. In addition, among all classifiers considered, KNN classifier achieved the highest classification accuracy on the test dataset.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The liver organ plays a critical role in many functions in the human body, including red blood cell decomposition. The liver is the largest organ in our body, and it is found in the upper right corner of our abdomen. Actually, any abnormality of the liver is referred to as liver disease. Some of the things that can happen with this disease are inflammation (hepatitis B and C) from infectious or non-infectious causes (chemical or autoimmune hepatitis), tumors, malignant scarring of the liver (cirrhosis), and metabolic disorders. Liver diseases can be caused by a variety of factors. High cholesterol, autoimmune disorders, and long-term use of medications are among them [1].
In the last few decades, liver diseases have rapidly increased in prevalence and severity, making them one of the leading global killers. According to the World Health Organization (WHO), chronic diseases are responsible for about 59% of global mortality and 46% of global diseases, and they kill about 35 million people every year [2]. Ultrasound (US), Computed Tomography (CT), and Magnetic Resonance Imaging (MRI) are just some of the imaging modalities used to diagnose liver disease [3, 4]. Nonetheless, it's possible that routine blood tests are essential in preventing liver disease [4]. The information age we are currently experiencing generates millions of data points daily from a wide variety of sources. Using machine learning techniques, these data can be used to enhance healthcare services and accurately diagnose diseases.
Researchers have paid a lot of focus to Machine Learning (ML), and it has been widely adopted and used in a wide variety of contexts around the world. In medicine, ML has demonstrated its effectiveness by being used to address a variety of urgent issues, including cancer treatment, heart disease diagnosis, dengue fever treatment, and other issues [5]. High data dimensionality is a problem that frequently arises in ML. Consequently, a large amount of memory is required, and sometimes these data may be irrelevant or redundant, resulting in an overfitting problem. Therefore, feature selection is carried out as a means of dealing with this issue. Feature selection is the process of deleting the less informative features and selecting the most informative ones [6].
Actually, it is too challenging to learn effective classifiers for many classification problems without first removing redundant features. It is possible to reduce the complexity of learning algorithms by removing superfluous features. There are numerous feature selection strategies available for determining which ones are the most informative. There are two types of these procedures: filter procedures and wrapper procedures [7,8,9]. Without a learning algorithm and instead using broad characteristics of the data, filter methods evaluate and select feature subsets. In contrast, wrapper approaches use a classification algorithm to assess a feature subset after an optimizing algorithm has been applied to either add or remove features [7,8,9]. Figures 1 and 2 depict respective examples of filter and wrapper techniques.
The primary contribution of this paper is the presentation of a new Liver Patients Detection Strategy (LPDS) that utilizes common blood tests for both Liver and non-Liver patients. In fact, the proposed LPDS is divided into three stages: (i) data preprocessing; (ii) feature selection; and (iii) detection. The primary goal of the data preprocessing phase is to eliminate any outliers from the input data using Isolation Forest (IF). Then, during feature selection phase, the most efficacious features are selected using Improved Binary Butterfly Optimization Algorithm (IB2OA) from the patient’s routine blood test to enable the detection phase to work well. In reality, IB2OA is a hybrid technique that incorporates both filter and wrapper strategies. There are two steps to IB2OA; the Primary Selection (PS) and the Final Selection (FS). In fact, PS is used to quickly identify the most important features. While FS is used to accurately select the most informative features. During detection phase, liver disease patients are detected based on the most efficacious features. Experimental results demonstrated that IB2OA outperforms other competitors as it introduces maximum accuracy.
Following this outline, the rest of the paper is presented as follows: Section 2 details the guiding principles of this piece. In Section 3, we will discuss the efforts made in the past to categorize patients with liver disease. The Liver Patients Detection Strategy (LPDS) is the topic of Section 4's extensive analysis. In Section 5, we discuss our findings from the experiments. In Section 6, conclusions are summarized.
2 Preliminary concepts
The principles used in this article include; swarm intelligence concept, butterfly optimization algorithm, and opposite based learning which are covered in detail in the following subsections.
2.1 Swarm intelligence
Swarm Intelligence (SI) is a method for modelling cooperative intelligence in biological systems. Natural swarm behavior is the basis for this well-liked multi-agent framework [10]. It acts similarly to how a pack of animals would in order to stay alive. Swarm behavior is advantageous for many kinds of animals in many different environments. According to models of basic group behavior, even relatively simple interactions between individuals can be enough to shape and display a variety of group morphologies. People form groups in order to share the burden of processing information and making business-related decisions. The group's ability to make better decisions than an individual does is known as "collective intelligence" [11, 12]. SI refers to the mechanism by which individuals interact with one another and the groups to which they belong [11, 12].
SI has been applied to the management of robots and unmanned vehicles, the forecasting of social behaviors, and the enhancement of computer and communication networks [10, 13, 14]. The field of SI algorithms is often thought of as a subset of AI. SI has recently attracted attention from the feature selection community [10, 13, 14] due to its ease of use and global search capabilities. Numerous algorithms that use swarm intelligence have been developed, such as Particle Swarm Optimization (PSO), Grey Wolf Optimizer (GWO), Genetic Algorithm (GA), Whale Optimizer Algorithm (WOA), Salp Swarm Algorithm (SSA), the Sine Cosine Algorithm (SCA), Bat Algorithm (BA), Ant Colony Optimization (ACO), and Butterfly Optimization Algorithm (BOA) [14, 15]. The search for the next iteration of any SI algorithm typically relies on a stochastic search algorithm, in which heuristic information is shared. The overarching structure of SI algorithms is depicted in Fig. 3.
It's essential to set the algorithm's parameters up front. Next, initialization and the accompanying strategies kick off the evolutionary process. In the next iteration of the SI framework, the fitness function will be used to rank the search agents. The fitness function can be either a simple metric, such as classification accuracy, or a more complex function. In a SI algorithm, the search agents are periodically updated and relocated in accordance with the algorithm's underlying mathematics. This procedure is carried out over and over again until the end condition is met. In the end, the optimal search result is found [14, 15].
2.2 Butterfly optimization algorithm
Butterfly Optimization Algorithm (BOA) is an example of a bio-inspired algorithm, a class of metaheuristic algorithms that takes inspiration from the natural world. Butterflies are used as search agents in the optimization process of BOA, which is based on their behavior when foraging for food [16, 17]. Butterflies are equipped with olfactory receptors that allow them to detect the aroma of food and flowers. Chemoreceptors are the sensory receptors that are found all throughout the butterfly's body. A butterfly is thought to be able to create scent or fragrance with some power or intensity in BOA [16, 17]. This scent has something to do with the butterfly's fitness, which is determined by the problem's objective function. This means that a butterfly's fitness will change as it moves from one position in the search space to another. The scent of a butterfly can be detected by other butterflies in the neighborhood, resulting in the formation of a collective social learning system. When a butterfly detects the scent of the best butterfly in the search space, it makes a beeline for it, and this stage is known as the BOA global search phase. While a butterfly in the search space is unable to detect the scent of another butterfly, this step is known as the local search phase since it will make random strides [16,17,18,19].
In BOA as in any SI algorithm, there are three phases which are; initialization phase, iteration phase, and final phase [20]. In the first phase (i.e., initialization phase), the parameters of the algorithm are defined after which, the algorithm generates its initial population randomly. Then, the algorithm enters the stage of iteration phase where the search agents use two phases which are; global search phase and local search phase according to a (p). switch probability Actually, p controls the algorithm strategy for both global and local searches [20]. The global and local searches of BOA, are determined using the following equations:
Where Yi(t + 1) represents the location of ith butterfly in the next repetition (t+1). Yi(t) donates the position of ith butterfly in the current repetition t, r is a random number between zero and one. In addition, Ybest(t) the location of the best butterfly in repetition t and Yj(t) and Yk(t) stand for jth and kth butterflies from the population in the current repetition t. f represents the perceived magnitude of the fragrance which is formulated by using the following equation:
Where c is the sensory modality, I is the stimulus intensity and a is the power exponent. In BOA equations (1) and (2) are used according to following strategy:
Where p is a constant number; p ∈ [0, 1]. Figure 4 shows the flow chart of convolutional BOA and conventional BOA algorithm is represented in algorithm 1.
2.3 Opposition-Based Learning
Opposition-Based Learning (OBL) is a new intelligent computing technology that has been successfully applied in many intelligent algorithm optimizations. OBL was proposed by Tizhoosh [21]. It has been theorized that OBL has a higher chance of finding a solution very close to the global optimal solution [22].
Definition 1: Opposite number
Let x a real number and yϵ[c, d], then the opposite number \(\overline{y}\) is given by the following equation:
Where \(\overline{y}\) is the opposite value y, of c and d refers to lower bound and upper bound respectively.
Definition 2: Opposite vector
If yi = {y1, y2, …. yn}; where y1, y2, …. yn are real numbers, then \({\overline{y}}_i\) is calculated using the following equation:
Let f(y) is the evaluation function, then in every repetition we calculate f(y) and \(f\left(\overline{y}\right)\) and the current solution will then be implemented by \(\overline{y}\) if \(f\left(\overline{y}\right)>f(y)\), otherwise y.
3 Related work
Previous efforts to categorize liver diseases were discussed in this section. An Image-based Classification Model (ICM) for liver disease classification has been proposed in [23] with novel methodology. The proposed method did go through several iterations. The used CT images were first preprocessed by cutting out the background and focusing in on the liver. Intensity and higher-order features were used to derive the 3D texture features. Then, we used a hybrid of the Whale Optimization Approach and Simulated Annealing (WOA-SA) to select the most effective features for further analysis. The proposed classification model is then fed with these informative features. The experimental results show that the proposed method outperforms the alternatives.
A machine learning-based Fatty Liver Disease Classification Model (FLDCM) is presented in [24]. Using four different classification techniques, the developed model successfully predicted fatty liver disease. Random Forest, Naive Bayes, Artificial Neural Networks, and Logistic Regression are the models in question. In the first stage of data preparation, all gaps in the data set were closed. The relative importance of each variable was then calculated using Information Gain (IG). Then, accurate FLD patient identification and predictive classification models were created. Compared to other classification models, the RF model performed better, as shown by the results in [24].
According to [25], a novel approach has been proposed to classify patients with liver cancer by using Serum Raman Spectroscopy in conjunction with Deep Learning Algorithms (SRS-DLA). Gaussian white noise was added to the data at levels of 5, 10, 15, 20, and 25 dB to increase the robustness of the proposed models. Convolutional neural networks that were fed data that had been enhanced by a factor of ten reportedly performed well, as shown in [25]. Using gadoxetic acid enhanced Hepatobiliary Phase (HBP) MRI, a fully automated Deep Learning (DL) algorithm has been introduced [26]. In the first step, convolutional neural network (CNN) input was generated from HBP images by creating representative liver patch images for each patient. Later on, the DL model was fed segmented liver images that had been patched together. As shown in [26], noninvasive liver fibrosis staging has good-to-excellent diagnostic performance in experimental settings.
Computer-assisted diagnosis (CAD) using ultrasound images of the liver has been developed, as detailed in [27]. The proposed CAD method used a voting-based classifier and machine learning algorithms to determine whether liver tissues were fatty or normal. First, a genetic algorithm was used to select multiple regions of interest (ROIs; a total of nine ROIs) within the liver tissue. Then, using Gray-Level Co-Occurrence Matrix (GLCM) and First-Order Statistics (FOS), 26 features of each ROI were extracted. Finally, fatty liver tissue was classified using a voting-based classifier. The results obtained in [27] show that the proposed CAD method outperformed the existing literature.
In [28], a novel framework for early detection of chronic liver disease is proposed. A model known as a Hybridized Rough Set and Bat-inspired Algorithm (HRS-BA) is being proposed. The primary goal of the proposed model is to give the doctor a new perspective. Decision-making factors were initially prioritized using BA. The decision rules were then created based on these characteristics. The results were also compared to those obtained by using hybridized decision tree algorithms, and they were found to be vastly superior. Current classification methods are briefly compared in Table 1.
4 The proposed Liver Patients Detection Strategy (LPDS)
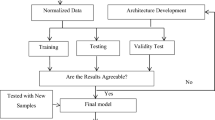
This section will go over the proposed Liver Patients Detection Strategy (LPDS). LPDS's primary goal is to detect patients who are infected with liver disease quickly and accurately. The early detection of liver disease patients allows for faster treatment and, consequently, slows the disease's spread. LPDS receives input in the form of a training set consisting of both normal and patient routine blood tests. After the model has been trained, it will be able to classify new cases. LPDS determines whether or not the input case is infected. As shown in Fig. 5, LPDS is divided into three phases: (i) data preprocessing, (ii) feature selection, and (iii) detection. The details of each phase will be discussed in the next subsection.
4.1 Preprocessing phase
The primary purpose of the preprocessing stage is to ready the data for the subsequent processing stage. Firstly, patient attributes are first extracted from the input training set. Table 2 lists several characteristics to consider when detecting liver disease patients. The dataset contains four missing values. However, special care must be taken with outliers. Two methods are employed. The results were unfavourable when machine learning algorithms were applied directly to the data without first removing outliers or selecting features. Results using the dataset's normal distribution to combat overfitting and then applying Isolation Forest (IS) for outlier detection are, however, quite encouraging [29, 30]. Several methods of plotting were used to examine the data for skewness, detect outliers, and verify the data's distribution. To succeed, each of these preprocessing methods is essential.
4.2 Feature selection phase
The most instructive features are chosen during the feature selection phase. The term "feature selection" describes this procedure. Feature selection is a method for increasing the classifier's accuracy by eliminating extraneous data points. Therefore, the feature selection process is crucial to enhancing the efficacy of learning algorithms [25, 31, 32]. Filter and wrapper methods are the two most common ways to categorize feature selection techniques [31, 32]. It has been demonstrated that filter methods are fast and scalable but cannot provide better performance than wrapper methods. However, wrapper methods are more expensive to compute [31,32,33], despite providing better performance. The Improved Binary Butterfly Optimization Algorithm (IB2OA) is proposed in this work as a new hybrid filter-wrapper approach to feature selection.
By combining the speed of the filter technique with the strength of the wrapper method, IB2OA is able to select features from the dataset with greater efficiency. For this reason, the proposed method seeks to simplify the calculations required to locate the optimal solution to high-dimensional datasets and cut down on the time spent doing so. Figure 6 shows that the core of the Improved Binary Butterfly Optimization Algorithm (IB2OA) is made up of two parts: (i) the Primary Selection (PS) using Information Gain (IG), and (ii) the Final Selection (FS) using IB2OA. The large search space slows down IB2OA's computation time, despite its ability to accurately identify the effective features. Therefore, the primary goal of PS is to apply IG to select the most effective features by narrowing the search space of B2OA, thereby reducing the time complexity. When all is said and done, the optimal subset of features helps enhance the reliability and performance of the employed classification model. Figure 6 shows how IG is used in PS to select the best possible set of useful and informative features to use when analyzing data from a dataset of liver patients.
Figure 6 depicts the extraction of features from a dataset of liver patients, followed by their transfer to the first step (e.g., PS) for preselection of the most effective and dynamic features. Therefore, the PS output will be fed into the second step (e.g., FS) to produce the seed "q" butterfly population. Then, IB2OA will be performed until the end point is reached. In the end, the most effective set of features is provided by the best solution for the population, IB2OA. Base classifiers, such as Naive Bayes (NB), should be used to assess this subset [34,35,36].
In general, IB2OA is relied on a meta-heuristic optimization algorithm called Butterfly Optimization Algorithm (BOA). BOA simulates the guidance and hunting behavior of the butterflies in search of food in natural environment. It was used to solve continuous optimization problem. Hence, to deal with feature selection problem which is considered discrete optimization problem, BOA is converted into B2OA. Hence, B2OA starts with a group of butterflies as solutions which are called Population (P). Each butterfly represents a candidate solution (e.g., the most efficacious subset of features). Each candidate solution represented as a binary vector of value ‘zero’ or ‘one’ in which its length is equal to the number of features. For features, a value of 'one' indicates selection, while a value of 'zero' indicates deselection or removal. Consequently, the size of the best subset is determined by a value of one’s.
Figure 6 shows the required sequential steps for implementing IB2AO as a feature selection. First, in PS, IG is used to preselect a set of optimum features from the extracted features from liver patients’ datasets. In other word, IG is used to rank the features based on its entropy using the following equation [37]:
Where Cn is the nth class category, f is stand for the feature. P(Cn) denotes the percentage of reviews in the Cn class category, and P(f) is the percentage of reviews in the f class category. \(P\left(\overline{f}\right)\) is the percentage of reviews that do not have the feature f. P(Cn| f) is the proportion of reviews in class category Cn that contain the feature f and \(P\left({C}_n|\overline{f}\right)\) is the proportion of reviews in class category Cnthat doesn’t contain the feature f.
Then, this subset of optimal features will be passed to the second step (e.g., FS) to be searched by using B2OA to determine the final best subset. As shown in Fig. 6, in FS, after generating the initial population of ‘q’ butterflies, EOBL is used to find the opposite solution for each initial solution. For example, if the initial solution y = {0, 1, 0, 1, 1, 0, 0, 1, 0, 1} thus, the opposite solution is \(\overline{y}=\left\{1,0,1,0,0,1,1,0,1,0\right\}\). Then, the whole solutions will be evaluated using the accuracy index of a standard classifier such as NB to find the best subset of features. for example, if the number of initial solutions is 20 solutions, then, the number of opposite solutions is also 20 solutions. Consequently, the final population contains 40 solutions and these solutions will be evaluated using the following equation:
Where Accuracy (Yi) is the success rate of classifying data using the ith set of features. The algorithm searches for the best butterfly with the highest Fit (Yi). After evaluating all candidate solution, the optimization procedure can be used to change the position of a butterfly that has been placed artificially using equation (4). After updating the new positions of the butterflies, these positions are position is adjusted using the sigmoid function, which is used to find new butterfly position relied on binary values by using (9):
Where \({Y}_i^j\left(t+1\right)\) represents the value of the ith butterfly at the jth position in the next repetition (t+1), where j=1,2, 3,...,m, and rand(0,1) represents a random number between [0,1]. Furthermore, the sigmoid transfer function \(\sigma \left({Y}_i^j\right)\) represents the likelihood that the jth bit is either 0 or 1. The formula for computing \(\sigma \left({Y}_i^j\right)\)
Each butterfly in P is scored using the fitness function in (8), which takes into account the butterfly's current position and its new position \({Y}_i^j\left(t+1\right)\). Then, the process is repeated up to the maximum allowed generations. Once the best butterfly has been selected from the population, the algorithm stops. All features contributed by 1 in this butterfly are the most reliable indicators of liver disease. Different features will be chosen as the best subset of features after the IB2OA algorithm is applied to the dataset of liver patients. Total Bilirubin (TB), Alkaline Phosphotase (Alkphos), Aspartate aminotransferase (Sgot), and the Albumin to Globulin Ratio (A/G Ratio) are the features chosen. Algorithm 2 depicts the proposed IB2OA's algorithm.
4.3 Detection phase
Finally, in the detection phase, different ML classifiers are used. Actually, the development in computer vision and ML technologies can be used for the accurate, quick, and earlier detection of liver disease patients [38]. Utilizing these technologies has the advantage of producing quick and precise results from computerized arrests. Time wastage can be decreased by utilizing improvements in computer vision and precision. Patients with liver disease benefit from being diagnosed early so that they can begin treatment as soon as possible. In this paper, the selected features are used to fed five different classifiers which are; Support Vector Machine (SVM), K- Nearest Neighbor (KNN), Naïve Bayes (NB), Decision Tree (DT), and Random Forest (RF) as shown in Fig. 7. Based on classification accuracy, the effectiveness of various classifiers was evaluated.
5 Experimental results
The effectiveness of the Liver Patients Detection Strategy (LPDS) that was just proposed will be discussed here. The proposed LPDS was used to identify infected patients with liver disease from laboratory results. In reality, there are three stages to LPDS: (i) data preprocessing, (ii) feature selection, and (iii) detection. During data preprocessing, the patient's information is managed, and anomalous data is eliminated. Following this, the most useful features are selected utilizing Improved Binary Butterfly Optimization Algorithm (IB2OA) during the feature selection phase. At last, these useful features are fed into five distinct classifiers: SVM, KNN, NB, DT, and RF. The most efficient of these classifiers will be selected on the basis of their ability to correctly categorize data. Our implementation is founded on a database of laboratory results for people with and without liver disease (https://www.kaggle.com/uciml/indian-liver-patient-records/home) [39]. With the help of the collected data (patients' dataset), the results presented in this paper were generated. Given the scarcity of publicly available datasets, the classification model is verified via cross-validation. Using 10-fold cross-validation, the dataset is split into 10 equal parts, with one part serving as the testing set and the other 9 as the training sets. As a result, there will be a total of 251 (90%) patients in the training phase and 28 (10%) patients in the testing phase. Our tests were run on an Intel Core i5-6200U processor at 2.30 GHz with 8 GB of RAM. Parameters and their corresponding values are shown in Table 3.
5.1 Data description
Information collected from patients' medical records is included in this dataset. Including the results of a battery of routine blood tests on people of varying ages, sexes, and health conditions (https://www.kaggle.com/uciml/indian-liver-patient-records/home). There are a total of 583 cases in the dataset. In fact, as shown in Table 4, the cases in the collected dataset are split into two groups: those with liver disease and those without. Hepatic patients are commonly referred to by that term. Patients who do not have liver disease are referred to as "non-liver patients." Ten features are used in both the training and testing datasets, all of which are derived from standard blood tests. Age, sex, Total Bilirubin (TB), Direct Bilirubin (DB), Alkaline phosphatase (Alkphos), alamine aminotransferase (Sgpt), aspartate aminotransferase (Sgot), Total Proteins (TP), albumin (ALB), and Albumin to Globulin ratio (A/G Ratio) (https://www.kaggle.com/uciml/indian-liver-patient-records/home) [39]. A breakdown of the collected data set by "Age," "Gender," and "Type of Disease" is shown in Figs. 8, 9 and 10.
5.2 Evaluation metrices
Measures of performance such as accuracy, error, precision, recall/sensitivity, and F-score are applied throughout this section to assess the quality of the proposed method [34,35,36]. The primary findings from the system used to assess the effectiveness of each algorithm are detailed in Tables 5 and 6. The confusion matrix is summarized using several different formulas in this paper, which are shown in Table 6 [34,35,36].
5.3 Comparison of IB2OA with standard Meta-heuristics
In this paper, a new feature selection methodology called Improved Binary Butterfly Optimization Algorithm (IB2OA) was introduced. Numerous feature selection techniques are compared to the proposed IB2OA based on the NB classifier as a base classifier in order to demonstrate the effectiveness of the proposed method. They include; Particle Swarm Optimization (PSO), Grey Wolf Optimizer (GWO), Genetic Algorithm (GA), Whale Optimizer Algorithm (WOA), Salp Swarm Algorithm (SSA), Sine Cosine Algorithm (SCA), Bat Algorithm (BA), Ant Colony Optimization (ACO), and Butterfly Optimization Algorithm (BOA). Table 7 displays the results.
According to Table 7, the proposed IB2OA performs better than other optimizers in terms of accuracy, precision, recall/sensitivity, and F-score. The proposed IB2OA achieved the highest accuracy of 0.988, as shown in Table 7, while the lowest value was 0.851, introduced by GWO. Furthermore, IB2OA introduces the highest precision value of 0.9009, while SCA introduces the lowest value of 0.699. IB2OA's average recall and F-score are, respectively, 0.8018 and 0.8902. Table 7 reveals that IB2OA performs significantly better than PSO, GWO, GA, WOA, SSA, SCA, BA, ACO, and BOA. The proposed strategy also showed steady behavior due to the STD values, as it obtained the lowest STD average across all methods.
5.4 Comparison with the state-of-the-arts FS methods
Through this subsection, the performance of the proposed method is compared to the state-of-the-art feature selection methods, including; Improved Binary Global Harmony Search (IBGSH) [40], Dynamic Salp Swarm Algorithm (DSSA) [41], Chimp Optimization Algorithm (ChOA) [42], Improved Binary Grey Wolf Optimizer (IBGWO) [43], Binary Monarch Butterfly Optimization (BMBO) [44], Information Gain binary Butterfly Optimization Algorithm (IG-bBOA) [17], and Enhanced Chaotic Crow Search and Particle Swarm Optimization Algorithm (ECCSPSOA) [45]. Table 8 displays the results.
Table 8 shows that the accuracy introduced by the proposed method is approximately 0.988, compared to for IBGSH, DSSA, ChOA, IBGWO, BMBO, IG-bBOA, and ECCSPOA, respectively. IB2OA achieved a precision of 0.8709 while IBGSH, DSSA, ChOA, IBGWO, BMBO, IG-bBOA, and ECCSPOA 's precisions are; 0.7821, 0.6802, 0.6920, 0.7397, 0.7063, 0.7318, and 0.8102 respectively. IB2OA 's recall value is 0.801 while DSSA's is 0.6755. Additionally, IB2OA provided the highest F-score value of 0.8302, while ChOA introduced the lowest value. Due to the STD values, which had the lowest average STD value when compared to the others, the proposed method also demonstrated stable behavior.
5.5 Testing the whole strategy
In this subsection, the whole strategy will be evaluated. In fact, five different classifiers which are; SVM, KNN, NB, DT, and RF will be tested to select the best one. Initially, the selected features are used to fed these classifiers. Based on the classification accuracy, the best one will be selected to complete the proposed strategy. The higher accuracy means the best classifier. Result is shown in Fig. 11.
Figure 11 introduces comparative study based on performance of five different classifiers. As shown in Fig. 11, the accuracy of the used of SVM, KNN, NB, DT, and RF are 96.2%, 99.1%, 98.8%, 93%, and 94.1 % respectively. From the results obtained, it is concluded that KNN give the best result with “K=5” as KNN is one of the most efficient ana simple algorithm used for disease classification. The encouraging results illustrate that the proposed IB2OA with KNN can accurately detect liver disease patients.
6 Conclusions
Chronic liver disease, the primary reason for death worldwide, affects a vast number of people. A variety of factors that harm the liver contribute to the development of this disease. For instance, alcohol abuse, undiagnosed hepatitis, and obesity which causes liver encephalopathy, jaundice, abnormal nerve function, bloody coughing or vomiting, kidney failure, liver failure, and many other symptoms. The diagnosis of this illness is very expensive and difficult. Therefore, it is essential to identify liver disease early in order to save lives and take the necessary steps to control the disease. This paper describes a new way to find people with liver disease. It is called Liver Patients Detection Strategy (LPDS). The proposed LPDS was depended on a new feature selection method called Improved Binary Butterfly Optimization Algorithm (IB2OA). IB2OA consists of two steps, which are Primary Selection Step (PS) and Final Selection Step (FS). Then, these features were used to feed five different classifiers, namely: Support Vector Machine (SVM), K-Nearest Neighbor (KNN), Naïve Bayes (NB), Decision Tree (DT), and Random Forest (RF) to detect liver patients. Results from experiments show that the proposed IB2OA has better average accuracy, average precision, average sensitivity/recall, and F-score than competing methods. Additionally, the proposed model's average selected features are 4.425 compared to the other state of the art. Additionally, KNN classifier has obtained the highest classification accuracy of 99.1% on test dataset among all classifiers taken into consideration.
Data availability
The dataset is available at: https://www.kaggle.com/uciml/indian-liver-patient-records/home
References
Karim M, Singal A, Kum H et al (2003) Clinical characteristics and outcomes of nonalcoholic fatty liver disease–associated hepatocellular carcinoma in the United States. Clin Gastroenterol Hepatol, Elsevier, 21(3):670-680
Moreau R, Tonon M, Krag A et al (2023) EASL Clinical Practice Guidelines on acute-on-chronic liver failure. J Hepatol, Elsevier 79(2):461–491
Hsu C, Caussy C, Imajo K et al (2019) Magnetic resonance vs transient elastography analysis of patients with nonalcoholic fatty liver disease: a systematic review and pooled analysis of individual participants. Clin Gastroenterol Hepatol, Elsevier 17:630–637. https://doi.org/10.1016/j.cgh.2018.05.059
Hydes T, Moore M, Stuart B et al (2021) Can routine blood tests be modelled to detect advanced liver disease in the community: model derivation and validation using UK primary and secondary care data. British Medical Journal (BMJ) 11(2):1–11. https://doi.org/10.1136/bmjopen-2020-044952
Amin R, Yasmin R, Ruhi S et al (2023) Prediction of chronic liver disease patients using integrated projection based statistical feature extraction with machine learning algorithms. Informatics in Medicine Unlocked, Elsevier 36:1–11. https://doi.org/10.1016/j.imu.2022.101155
Houssein E, Hosney M, Mohamed W et al (2023) Fuzzy-based hunger games search algorithm for global optimization and feature selection using medical data. Neural Computing and Application, Springer 35:5251–5275. https://doi.org/10.1007/s00521-022-07916-9
Shaban W (2023) Insight into breast cancer detection: new hybrid feature selection method. Neural Computing and Applications, Springer 35:6831–6853. https://doi.org/10.1007/s00521-022-08062-y
Singh N, Singh P (2021) A hybrid ensemble-filter wrapper feature selection approach for medical data classification. Chemometrics and Intelligent Laboratory Systems, Elsevier, 217, https://doi.org/10.1016/j.chemolab.2021.104396
Mandal M, Singh P, Ijaz M et al (2021) A tri-stage wrapper-filter feature selection framework for disease classification. Sensors, Multidisciplinary Digital Publishing Institute (MDPI) 21(16):1–24
Tang J, Duan H, Lao S (2023) Swarm intelligence algorithms for multiple unmanned aerial vehicles collaboration: a comprehensive review. Artificial Intelligence Review, Springer 56:4295–4327. https://doi.org/10.1007/s10462-022-10281-7
Tang J, Liu G, Pan Q (2021) A review on representative swarm intelligence algorithms for solving optimization problems: applications and trends. Journal of Automatica Sinica, IEEE 8(10):1627–1643
Brezocnik L, Fister I Jr, Podgorelec V (2018) Swarm intelligence algorithms for feature selection: a review. Applied Science, Multidisciplinary Digital Publishing Institute (MDPI) 8:1–31. https://doi.org/10.3390/app8091521
Xue J, Shen B (2020) A novel swarm intelligence optimization approach: sparrow search algorithm. Systems Science & Control Engineering, Taylor & Francis 8(5):22–34
Nguyen B, Xue B, Zhang M (2020) A survey on swarm intelligence approaches to feature selection in data mining. Swarm and Evolutionary Computation, Elsevier 54:1–27. https://doi.org/10.1016/j.swevo.2020.100663
Kicska G, Kiss A (2021) Comparing swarm intelligence algorithms for dimension reduction in machine learning. Big Data and Cognitive and Computing, Multidisciplinary Digital Publishing Institute (MDPI) 5(3):1–15
Alweshah M, Al Khalaileh S, Gupta B et al (2020) The monarch butterfly optimization algorithm for solving feature selection problems,” Neural Computing and Applications, Springer, pp. 1-15 https://doi.org/10.1007/s00521-020-05210-0
Sadeghian Z, Akbari E, Nematzadeh H (2021) A hybrid feature selection method based on information theory and binary butterfly optimization algorithm. Engineering Applications of Artificial Intelligence, Elsevier 97:1–13. https://doi.org/10.1016/j.engappai.2020.104079
Long W, Jiao J, Liang X et al (2021) Pinhole-imaging-based learning butterfly optimization algorithm for global optimization and feature selection. Applied Soft Computing, Elsevier 103:1–19. https://doi.org/10.1016/j.asoc.2021.107146
EL-Hasnony I, Elhoseny M, Tarek Z (2022) A hybrid feature selection model based on butterfly optimization algorithm: COVID-19 as a case study. Expert Systems, Wiley Online Library 39:1–28. https://doi.org/10.1111/exsy.12786
Thawkar Sh, Sharma S, Khanna M et al (2021) Breast cancer prediction using a hybrid method based on Butterfly Optimization Algorithm and Ant Lion Optimizer. Computers in Biology and Medicine, Elsevier, 139, https://doi.org/10.1016/j.compbiomed.2021.104968
Tizhoosh H (2005) Opposition-based learning: a new scheme for machine intelligence. In: Proceedings of the International Conference on Computational Intelligence for Modelling, Control and Automation (CIMCA ’05) and International Conference on Intelligent Agents, Web Technologies and Internet Commerce (IAWTIC ’05), pp. 695–701
Hussien A, Amin M (2022) A self-adaptive Harris Hawks optimization algorithm with opposition-based learning and chaotic local search strategy for global optimization and feature selection. International Journal of Machine Learning and Cybernetics, Springer 13:309–336. https://doi.org/10.1007/s13042-021-01326-4
Rajathi G, Jiji G (2019) Chronic Liver Disease Classification Using Hybrid Whale Optimization with Simulated Annealing and Ensemble Classifier. Symmetry, Multidisciplinary Digital Publishing Institute (MDPI) 11(33):1–21
Wua C, Yehb W, Hsu W et al (2019) Prediction of fatty liver disease using machine learning algorithms. Computer Methods and Programs in Biomedicine, Elsevier 170:23–29. https://doi.org/10.1016/j.cmpb.2018.12.032
Meng C, Li H, Ch C et al (2022) Serum Raman spectroscopy combined with Gaussian—convolutional neural network models to quickly detect liver cancer patients. Spectroscopy Letters, Taylor & Francis Online 55(2):79–90
Hectors S, Kennedy P, Huang K et al (2021) Fully automated prediction of liver fibrosis using deep learning analysis of gadoxetic acid–enhanced MRI. European Radiology, Springer 31:3805–3814. https://doi.org/10.1007/s00330-020-07475-4
Gaber A, Youness H, Hamdy A et al (2022) Automatic classification of fatty liver disease based on supervised learning and genetic algorithm. Appl Sci, MDPI 12(1):1-15
Acharjya D, Ahmed P (2022) A hybridized rough set and bat-inspired algorithm for knowledge inferencing in the diagnosis of chronic liver disease. Multimedia Tools and Application, Springer:1–24. https://doi.org/10.1007/s11042-021-11495-7
Heigl M, Anand K, Urmann A et al (2021) On the Improvement of the Isolation Forest Algorithm for Outlier Detection with Streaming Data. Electronics, Multidisciplinary Digital Publishing Institute (MDPI) 10:1–26. https://doi.org/10.3390/electronics10131534
Zhang S, Carranza M, Xiao K et al (2022) Mineral Prospectivity Mapping based on Isolation Forest and Random Forest: Implication for the Existence of Spatial Signature of Mineralization in Outliers. Natural Resources Research 31:1981–1999. https://doi.org/10.1007/s11053-021-09872-y
Pramanik R, Pramanik P, Sarkar R (2023) Breast cancer detection in thermograms using a hybrid of GA and GWO based deep feature selection method. Expert Systems with Applications, Elsevier 219:1–12. https://doi.org/10.1016/j.eswa.2023.119643
Bharti R, Khamparia A, Shabaz M et al (2021) Prediction of Heart Disease Using a Combination of Machine Learning and Deep Learning. Computational Intelligence and Neuroscience, Hindawi 2021:1–11. https://doi.org/10.1155/2021/8387680
Sathiyabhama B, Kumar S, Jayanthi J et al (2021) A novel feature selection framework based on grey wolf optimizer for mammogram image analysis. Neural Computing and Applications, Springer:1–20. https://doi.org/10.1007/s00521-021-06099-z
Shaban W, Rabie A, Saleh A et al (2021) Detecting COVID-19 patients based on fuzzy inference engine and Deep Neural Network. Applied Soft Computing, Elsevier 99:1–19. https://doi.org/10.1016/j.asoc.2020.106906
Shaban W, Rabie A, Saleh A et al (2020) A new COVID-19 Patients Detection Strategy (CPDS) based on hybrid feature selection and enhanced KNN classifier. Knowledge- Based Systems, Elsevier 205:1–18. https://doi.org/10.1016/j.knosys.2020.106270
Shaban W, Rabie A, Saleh A et al (2021) Accurate detection of COVID-19 patients based on distance biased Naïve Bayes (DBNB) classification strategy. Pattern Recognition, Elsevier 119:1–15. https://doi.org/10.1016/j.patcog.2021.108110
Zhang G, Hou J, Wang J et al (2020) Feature Selection for Microarray Data Classification Using Hybrid Information Gain and a Modified Binary Krill Herd Algorithm. Interdisciplinary Sciences: Computational Life Sciences, Springer 12:288–301. https://doi.org/10.1007/s12539-020-00372-w
Gumbs A, Grasso V, Bourdel N et al The advances in computer vision that are enabling more autonomous actions in surgery: a systematic review of the literature. Sensors, Multidisciplinary Digital Publishing Institute (MDPI) 22:1–21. https://doi.org/10.3390/s22134918
Singha J, Baggab S, Kaur R (2020) Software-based Prediction of Liver Disease with Feature Selection and Classification Techniques. Procedia Computer Science, Elsevier 167:1970–1980. https://doi.org/10.1016/j.procs.2020.03.226
Gholami J, Pourpanah F, Wang X (2020) Feature Selection based on Improved Binary Global Harmony Search for Data Classification. Applied Soft Computing, Elsevier 93:1–20. https://doi.org/10.1016/j.asoc.2020.106402
Tubishat M, Ja’afar S, Alswaitti M et al (2021) Dynamic Salp swarm algorithm for feature selection. Expert Systems with Applications, Elsevier 164:1–15. https://doi.org/10.1016/j.eswa.2020.113873
Pashaei E, Pashaei E (2022) An efficient binary chimp optimization algorithm for feature selection in biomedical data classification. Neural Computing and Applications, Springer 34:6427–6451. https://doi.org/10.1007/s00521-021-06775-0
Hu P, Shyang J, Chua P (2020) Improved Binary Grey Wolf Optimizer and Its application for feature selection. Knowledge- Based Systems, Elsevier, 195:1-14. https://doi.org/10.1016/j.knosys.2020.105746
Sun L, Si S, Zhao J et al (2022) Feature selection using binary monarch butterfly optimization. Appl Intell, Springer, pp. 1-22. https://doi.org/10.1007/s10489-022-03554-9
Adamu A, Abdullahi M, Junaidu S et al (2021) An hybrid particle swarm optimization with crow search algorithm for feature selection. Machine Learning with Applications, Elsevier, 6, pp. 1-13 https://doi.org/10.1016/j.mlwa.2021.100108
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shaban, W.M. Early diagnosis of liver disease using improved binary butterfly optimization and machine learning algorithms. Multimed Tools Appl 83, 30867–30895 (2024). https://doi.org/10.1007/s11042-023-16686-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-16686-y