Abstract
In the western world,the prostate cancer is major cause of death in males. Magnetic Resonance Imaging (MRI) is widely used for the detection of prostate cancer due to which it is an open area of research. The proposed method uses deep learning framework for the detection of prostate cancer using the concept of Gleason grading of the historical images. A3D convolutional neural network has been used to observe the affected region and predicting the affected region with the help of Epithelial and the Gleason grading network. The proposed model has performed the state-of-art while detecting epithelial and the Gleason score simultaneously. The performance has been measured by considering all the slices of MRI, volumes of MRI with the test fold, and segmenting prostate cancer with help of Endorectal Coil for collecting the images of MRI of the prostate 3D CNN network. Experimentally, it was observed that the proposed deep learning approach has achieved overall specificity of 85% with an accuracy of 87% and sensitivity 89% over the patient-level for the different targeted MRI images of the challenge of the SPIE-AAPM-NCI Prostate dataset.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The prostate cancer refers to the enlargement of the prostate gland in the reproductive system of males. Mostly, prostate cancer grows slowly, however, sometimes it can enlarge quickly [1]. The infected cells can also spread from cancerous areas to another unaffected cell mainly to the lymph nodes or the bones which shows no symptoms in its initial stage [2]. In further stages, there might be difficulties while urinating, blood in the urine, pelvis, and back pain, and its further stages of symptoms we can feel tired because of the minimum level of red blood cells.
In the American cancer society, it was observed that around 174,750 men were diagnosis in 2019 for having prostate cancer. Currently, around 3.1 million men in America were having these diseases which aresimilar to the population of Chicago [3]. Early treatment is only the way for live-saving of the patients with prostate cancer for the long term and the free survival progression [10]. Recently, it was observed that the survival rate of men having prostate cancer is close to 100% for the last 10 years [18].
Prostate cancer treatment may include continuous surveillance and many therapies like radiation, externalbeam, and proton therapy. Its treatment also includes cryosurgery ultrasound of high intensity and chemotherapy as well as hormonal therapy and at last stage prostatectomy treatment with the radiation therapy [19].
The system of grading the Gleason helps in evaluating the growth of the prostate cancer from the sample collected from prostate biopsy. This helps in detecting the prognosis and guide in further treatment of prostate cancer [20]. The microscope appearance of the infected cells helps in generating the Gleason score which helps in determining the cancer stage with its prognosis [22]. The Gleason grade obtained from microscopic helps to form the dominant and morphological pattern with its grade which collectively generates the result of having a biopsy of normal and the abnormal tissue grading from 1 to 10 as the pattern of biopsy generates [23, 30].
Magnetic Resonance Imagining (MRI) has shown significant improvement while visualizing prostate cancer and helps in targeting the biopsies of the prostate which visualizes the infected area in MRI [14]. Biopsy as Transrectal Ultrasound (TRUS) and the biopsy of the Transperineal Template (TPM) were used systematically for detecting the cancerous cell [11]. Biomarkers of prostate cancer have an extended range for their biochemical entities in the form of protein, urine, and nucleic acid. Biomarkers are mostly used for evaluating the risk assessments, help in diagnosing and investigating the infected cells, and helps in predicting the accurate therapy for its treatment [13]. MRI-TB defines the index testing and guides in transrectal biopsy which identifies the affected region in the MRI image. Magnetic resonance imaging-guided targeted biopsy helps to generate the positive PSA test [4]. Diffusion-Weighted Imaging (DWI) and the Dynamic Contrast-Enhanced (DCE) play a significant role in measuring the water molecules present in tissue through Brownian motion which depicts the vascular and the vascularity of tissue permeabilityover the different time through different signal [27].
The MRI and MRI-TB make alternatives while diagnosing PC as systematic biopsy [17]. Randomized Controlled Trials (RCTs) were considered with MRI-TB and systematic biopsy. Besides, an analysis of post hoc sensitivity was performed. This entirely enhanced the PCa clinical suspicion where MRI-TB shows more efficient results than a traditional systematic biopsy and needs fewer biopsy cores for diagnosing the PC patients. Prostate Imaging Reporting and Data System (PI-RADS) [12] were used with the multiparametric MRI for detecting prostate cancer. They analyze PI-RADS accuracy with mp-MRI where they searched for the Medline and the Embase using biopsy or prostatectomy as analyzed by two different independent reviewers. Data of 1785 patients were analyzed having 0.78 sensitivity, witha confidence interval of 95% for PC detection. PC-RADS criteria are used as it has good PC detection accuracy. The recurrent network for detection of PC [16] using an antigen of the specific membrane having Tomography emission of positron which doesn’t phoenix criteria observed in curative radiography.
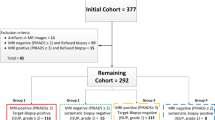
Radiography defines PSA(Prostate-Specific Antigen). Here, PSMA-PET scanned were taken from 315 patients some have phoenix threshold below which is observed importantly [5]. Some patients were treated with Brachy Therapy (BT) or External-Beam Radio Therapy (EBRT) for finding PC which was generated by Biochemical Recurrence (BCR) of disease. PCa detection with this recurrence radiotherapy has shown a rise in PSA value below the phoenix threshold of PSA having 0.2 ng/ml which is above nadir. Again, the entire data plea for re-evaluation for the current state of PSA value rising after radiography which improves selection of patients for salvage or directed treatment of oligometastatic [28]. Prostate Cancer is classified according to the Gleason grade score which ranges from one to five by combining two different groups and finally scores from one to five [6].The data pre-processing pipeline is shown in Fig. 1.
Pipeline for data preparation concerning false-negative lesions and mp-MRI [4]
2 Data acquisition
2.1 Objectives
The accuracy of the diagnostic was determined based on MRI tests, a biopsy of MRI, pathway MRI, and the systematic biopsy when it is compared with guided-template biopsy as standard marked by the International Society of Urological Pathology (ISUP) in its Gleason score of grades 2 or even higher condition of the target [12]. Furthermore, we focus on determining its agreement or disagreement in the change in the potential biopsy number which is collected from two different index tests and the pathway of MRI and the systematic biopsy for determining the affected region in the MRI images [6]. Prostate Imaging Reporting and Data System (PI-RADS) have reported the cohort subgroup when compared with prostate-specific antigen level.
2.2 TRUS/MRI fusion biopsy
The MRI biopsy fusion system was commonly used for mostly biopsies. Ginsburg protocol state the volume of the prostate for directed grid transperineal biopsy sector with the standard technique of anaesthesia [7]. Cores of four TB median were obtained from every prior lesion to perform biopsy transperineal fusion for the visualization of TRUS. TMP or the TRUS biopsy helps to define systematic biopsy. An insignificant clinical definition was not accepted for the determination of cancer further the Gleason grade was used for predicting cancer. Prostate Biopsy [26] wasconduct for gaining the information of multi-parametric MRI biopsy which states the MRI-TB.
2.3 Study population
The population study consisted of different men having clinical suspicionof having prostate cancer which is classified based on prostate-specific antigen and the examination of rectal digitally in the form of biopsy-naïve [29]. Biopsy template of guided mapping has transperineal which served them as a standard reference for conforming target of biopsy histopathological as mentioned by Standard of Reporting for MRI-targeted biopsy studies (START) which overcomes the clarity between methods of biopsy and its definition.
3 Methods
3.1 Data
The dataset consists of 346 patients consecutively with the score of PI-RADS of 3 or even more.All the different studies indicate that T2-Weighted (T2W), Dynamic Contrast-Enhanced (DCE), Proton Density-Weighted (PD-W), and the imaging of Diffusion-Weight (DW). The DICOM images comprise several series each comprising several instances. Each patient has one study with several DICOM images and one Ktrans images. Out of 349 studies and the 346 patients,112 findings (lesion) were present in the training set while there are 70 findings (lesion) in the testing set [21]. Data dataset contains total of 309,251 images are having a size of 15.1 GB with total of 18,321 number of series are present [8].
The different location of the lesion was shown in different excel format having their truth level with the centroid of lesion which shows the Gleason Grade respectively.Here, we visualize the Gleason score higher or more than 7 as given by the International Society of Uropathology with a Gleason Grade (GG > =3) as the patient has prostate cancer and below these patientsdoesn’t have cancer. The Data used in this study shown in Table 1.
Each MRI cases includes various set of MRI series, out of which only 56 MRI images belongs to four sets of MRI scan data is used for this study. 32 MRI images of T2-weighted images (transaxial and sagittal) available in DICOM format, 12 Ktrans images, computed from dynamic contrast-enhanced (DCE) images, available in. mhdand. Zraw format and 12 apparent diffusion coefficient (ADC) images (computed from diffusion-weighted (DWI) imaging) available in DICOM format are used for this study.
3.2 MRI acquisition
Two different types of Siemens a 3 T MR scanner, namely MAGNETOM Trio and Skyra has been used for acquire MRI images. Prostate cancer was diagnosis by the image of T2-weighted which contains nearly two different functional modalities obtained from the images of DCE, DWI, and the spectroscopic. Images of T2-weighted were obtained using the sequence of turbo spin-echo which has a resolution of 0.5 mm in the plane and 3.6 mm thickness of the slice. The series of DCE time were obtained by using the resolution of 1.5 mm round in-plane and the 4 mm slice thickness with the 3.5 s of temporal resolution. Finally, the series of DWI were obtained with help of a sequence of single-shot echo-planar having the 2 mm resolution in-plane and the thickness of 3.6 mm with the grading diffusion-encoding in three different areas. The ground truth images are marked by expert radiologist, with more than 20 years of experience in prostate MR. Figure 2 shows MR images and ground truth marked image available in dataset.
3.3 Methodology outlines
The methodology used in this paper includes processes in the following order: convert Digital Imaging and Communications in Medicine (DICOM) images to .png or .jpeg format, read multiple MRI images, stack images to get a sense of image in 3D, data augmentation, image pre-processing, transfer learning using TensorFlow Object Detection APIs such as inception ResNet-v2 [24]. During training, some extra steps such as cropping images to focus on affected regions, locating coordinates of affection regions and place, reading labels, and processing them, and then finally dumping the final dataset into a pickle file for further training and testing was applied. A pickle file is used to convert a python object hierarchy to byte stream. It is used in python programming language to store the python intermediate results. The dataset was dumped into pickle format for simplicity of loading different data instances and types obtained from different pre-processing techniques and data augmentation processes. The steps can be visualized in Fig. 3.
3.4 Data pre-processing and augmentation
The Pre-processing involved in this research starts from the input of the DICOM format file since MRI scans are usually distributed in this format. The DICOM file contains multiple MRI images along with metadata of patients in which sensitive information was already excluded from the dataset.During this research, the metadata was not taken into consideration so was not useful and removed from further processing. Further, the dataset was very large and contained various noises which could easily lead to overfitting of the model. To prevent this data filtered and various noisy data was removed. The DICOM files were then loaded and converted into Neuroimaging Informatics Technology Initiative (NIfTI) file format for their usability in the data processing. The NIfTI file format was then converted to png or jpeg format for deep learning models.
After this, the images were stacked together (each MRI scan event has a single data point) for every patient.The major data augmentation that took place was resizing the images into the shape of 32 × 32 along with other many augmentation processes. The data augmentation is applied with random zoom, random rotation [±15°, ±30°, ±45°, ±60°, ±90°], random flip along horizontal & vertical on training data.
Convolutional Neural Network (CNN) is a special Neural Network that is specifically optimized for images and hence explicitly assumes the input to be imaged [9]. CNN contains various layers of convolution layers, pooling layers, and filtering layers all for images to be properly processed and learned from. These layers help a deep learning network to filter images and process them properly according to need and tune the parameters (learnable parameters) accordingly to perform learning. The TensorFlow Object Detection models used in this research have the same basic CNN but have been upgraded and optimized greatly to specifically perform object detection. The Faster RCNN Inception v2 trained on the COCO dataset is a great model for objection detection purposes. Regional CNN (RCNN) are the same as regular CNNs except that the goal of RCNNs is to take an input image and output object detection using bounding boxes around the detected objects, as a result, RCNNs have been modified accordingly. In the earlier days, RCNNs used to apply a method of Selective Selection to produce multiple, possibly thousands of Regions of Interest (ROI) [19]. This ROI are the bounding regions/rectangles focusing on a specific region that may represent an object inside. Each ROI was then fed to a neural network to produce an output feature which was used to determine if the region contains anyobject using Support Vector Machine classifiers [25].Spectral clustering approach with Gaussian similarity has been used [15] to demark the prostate boundaries. The author has achieved segmentation accuracy of 92% on MR images. The proposed 3D CNN architecture for this can be shown in Fig. 4.
4 Result and discussion
4.1 Experimental setup
All the experiments have been conducted on personal computer having the NVIDIA GeForce RTX 2060 with 4GB graphics and the processor of core i7 10th gen with an SSD of 500GB and the 16GB DDR4 ram with the variant of the processor of 10750H having the clock speed of 2.6 GHz with the turbo boost up to 5.0GHz with the ram frequency of 2666 MHz. Python v3 language has used to implement and training of the deep learning models.
4.2 Results and discussion
The research can be divided into two parts, pre-processing, and training along with testing. The pre-processing required various Python packages like pydicom (for reading and processing DICOM files), OpenCV(image processing library), DICOM2NIFTI (to convert DICOM files to NIfTI format), Dipy (an s/w project for computational neuroanatomy mainly for diffusion Magnetic Resonance Imaging (dMRI) images), Pickle (for dumping files in pickle format and loading them) along with other basic python package tools. Followed by pre-processing are the training and testing phase. For this, the labels also need to be properly organized and processed. For testing and training, TensorFlow Object Detection API was installed on the working machine according to the official documentation. After it was working and running the data dumped on a pickle file was loaded and prepared to be fed into an object detection model. The training took a relatively long time but due to GPU availability on Google Colab, it was easier and faster. After the models were properly trained, the model was exported as a frozen inference graph which was to be loaded during testing or for making predictions.
The process explained above can be broken down into simpler steps. First of all, the input data was loaded, which was in the form of DICOM file format. These files were loaded(extracted) into the system and were converted into NIfTI file format. Since the data was in large amount and divided into different folders for each test of each session of every patient, a directory crawler was developed to crawl an input data directory to find, load, convert to NIfTI, and export/save every DICOM file to a similarly structurally created directories, also done by the same program. After all, DICOM files are converted into NIfTI format, these NIfTI files can either be saved on the directory or further processed to export png or jpeg format images. The images saved on the output directory are loaded onto the system to perform further pre-processing and data augmentation as shown in Fig. 5.
The images were stacked to make sense of them on a 3D surface. The labels were read, properly processed, and prepared as input to the deep learning model. During this, the image pre-processing and augmentation processes like filtering, blur, crop, etc. can differ during research to produce the best results as shown in Fig. 6. Hence, each instance of data obtained after a certain process was loaded and dumped on a pickle file. These data instances can be loaded to train first and later test the models to observe the performance of the overall process. After dumping the data in a pickle file, the pickle file can be loaded into the training and testing system where TensorFlow Object Detection API is properly installed and running. Inceptio-ResNet-V2 is pre-trained neural network, which is trained on large scale image classification ImageNet dataset. FasterRCNN receives two inputs one from region proposed by features extracted using Inception-ResNet-V2 and another input image. Faster RCNN provide the out object surrounded by bounding box or region with maximum probability. The training and testing for segmentation has been implemented using FasterRCNN with Inception-ResNet-V2 object detection models and results can be visualized in Fig. 7. The processes involved in installing, training, and testing the object detection models on the API can be found on its official documentation of TensorFlow.
The confusion matrix was visualized on validation data for generating performance of proposed model for predicting cancer. The confusion matrix is generated for visualizing the performance of the model by plotting the true negative rates and the true positive rates obtained by using different thresholds. For binary classification task performance metrics such as accuracy, precision, sensitivity, specificity and F1-score can be calculated using following formula.
As discussed in proposed methodology in Fig. 4, 32 ADC MR Images of size 32 × 32, 12 high b-Value MR images of same size 32 × 32 and 12 Ktrans MR images of size 32 × 32 are stacked for each patient. The result indicates that region of lesion is positive, or negative are difficult to predict. The stack of image is said to be positive when it visualized white color higher than grey, and negative is referred when grey color is higher than white. The patient wise output confusion matrix using proposed method is shown in Fig. 8.
The test datasets contain the MRI images of 142 patients, which includes 70 findings (lesion) and 72 non-lesions. Performance metrics such as Accuracy, Precision, recall, F1-Score and Specificity can be calculated and the values are shown in Table 2.
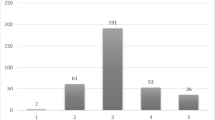
From Table 2 we can observe that Ktrans images improve the classification accuracy. It can also be observed that proposed method with 52 stacked MR images of ADC, BVAL and Ktrans improve the overall performances. Performance comparison in bar chart is shown in Fig. 9. Performance comparison of the proposed algorithm with state-of-art methods shown in Table 3.
5 Conclusion and future work
The suggested methodology can classify prostate cancer which belongs to the separate group of Gleason grade having a different quadratic weight of the cancerous score. The proposed technique helps in predicting prostate cancer significantly. The final result was obtained using the prostate-2 dataset which gives a better result than other existing methodology with proposed CNN. The overall accuracy achieved is up to 87%, specificity 85% and sensitivity 89%. The proposed method also shows segmented result of lesion detected using Faster RCNN with Inception-Resnet-V2 transfer learning methods.
Even though results are better, further many experiments using the dataset for obtaining results more efficiently. For its clinical detection, the proposed methodology needs some more improvement for getting better accuracy. Other fields in potential improvement can be the layer of “ROIAlign”. The layer of “ROIAlign” generates the tiny feature map from its corresponding pyramid layer feature obtained from each ROI before the head of the network which results in degradation of some scale information that could be relevant while collecting information for histopathology.
References
Artan Y, Yetik IS (2012) Prostate cancer localization using multiparametric MRI based on semisupervised techniques with automated seed initialization. IEEE Trans Inf Technol Biomed 16(6):1313–1323. https://doi.org/10.1109/TITB.2012.2201731
Azizi S, Bayat S, Yan P, Tahmasebi A, Kwak JT, Xu S, Turkbey B, Choyke P, Pinto P, Wood B, Mousavi P (2018) Deep recurrent neural networks for prostate cancer detection: analysis of temporal enhanced ultrasound. IEEE Trans Med Imaging 37(12):2695–2703. https://doi.org/10.1109/TMI.2018.2849959
Brunese L, Mercaldo F, Reginelli A, Santone A (2019) Prostate Gleason score detection and cancer treatment through real-time formal verification. IEEE Access 7:186236–186246. https://doi.org/10.1109/ACCESS.2019.2961754
Cao R, Bajgiran AM, Mirak SA, Shakeri S, Zhong X, Enzmann D, Raman S, Sung K (2019) Joint prostate Cancer detection and Gleason score prediction in mp-MRI via FocalNet. IEEE Trans Med Imaging 38(11):2496–2506. https://doi.org/10.1109/TMI.2019.2901928
Chen LC, Papandreou G, Kokkinos I, Murphy K, Yuille AL (2018) Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans Pattern Anal Mach Intell 40(4):834–848
Cheng R, Roth HR, Lu L, Wang S, Turkbey B, Gandler W, McCreedy ES, Agarwal HK, Choyke P, Summers RM, McAuliffe MJ (2016) “Active appearance model and deep learning for more accurate prostate segmentationon MRI”, In: Medical Imaging 2016: Image Process 9784. https://doi.org/10.1117/12.2216286
Cheng R, Roth HR, Lay N, Lu L, Turkbey B, Gandler W, McCreedy ES, Pohida T, Pinto PA, Choyke P, McAuliffe MJ, Summers RM (2017) Automatic magnetic resonance prostate segmentation by deep learning with holistically nested networks. J Med Imaging (Bellingham) 4(4):041302. https://doi.org/10.1117/1.JMI.4.4.041302
Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L, Prior F (2013) The Cancer imaging archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 26(6):1045–1057. https://doi.org/10.1007/s10278-013-9622-7
Dai Z, Carver E, Liu C, Lee J, Feldman A, Zong W, Pantelic M, Elshaikh M, Wen N (2020) Segmentation of the prostatic gland and the intraprostatic lesions on multiparametic magnetic resonance imaging using mask region-based convolutional neural networks. Adv Radiat Oncol 5(3):473–481
Feng Y, Yang F, Zhou X, Guo Y, Tang F, Ren F, Guo J, Ji S (2019) A deep learning approach for targeted contrast-enhanced ultrasound based prostate Cancer detection. IEEE/ACM Trans Comput Biol Bioinforma 16(6):1794–1801. https://doi.org/10.1109/TCBB.2018.2835444
Gorelick L, Veksler O, Gaed M, Gomez JA, Moussa M, Bauman G, Fenster A, Ward AD (2013) Prostate histopathology: learning tissue component histograms for cancer detection and classification. IEEE Trans Med Imaging 32(10):1804–1818. https://doi.org/10.1109/TMI.2013.2265334
Hamoen EHJ, De Rooij M, Witjes JA, Barentsz JO, Rovers MM (2015) Use of the prostate imaging reporting and data system (PI-RADS) for prostate cancer detection with multiparametric magnetic resonance imaging: a diagnostic meta-analysis. Eur Urol 67(6):1112–1121. https://doi.org/10.1016/j.eururo.2014.10.033
Hassanzadeh T, Hamey LGC, Ho-Shon K (2019) Convolutional Neural Networks for Prostate Magnetic Resonance Image Segmentation. IEEE Access 7(c):36748–36760. https://doi.org/10.1109/ACCESS.2019.2903284
Hussain L, Ahmed A, Saeed S, Rathore S, Awan IA, Shah SA, Majid A, Idris A, Awan AA (2018) Prostate cancer detection using machine learning techniques by employing combination of features extracting strategies. Cancer Biomark 21(2):393–413. https://doi.org/10.3233/CBM-170643
Ingale K, Shingare P, Mahajan M (2023) Spectral clustering to detect malignant prostate using multimodal images, ICDSMLA 2021: proceedings of the 3rd international conference on data science, machine learning and applications. Springer Nature Singapore, Singapore
Jansen BH, van Leeuwen PJ, Wondergem M, van der Sluis TM, Nieuwenhuijzen JA, Knol RJ, van Moorselaar RJ, van der Poel HG, Oprea-Lager DE, Vis AN (2020) Detection of recurrent prostate Cancer using prostate-specific membrane antigen positron emission tomography in patients not meeting the Phoenix criteria for biochemical recurrence after curative radiotherapy. Eur Urol Oncol 2005:1–5. https://doi.org/10.1016/j.euo.2020.01.002
Kasivisvanathan V, Stabile A, Neves JB, Giganti F, Valerio M, Shanmugabavan Y, Clement KD, Sarkar D, Philippou Y, Thurtle D, Deeks J (2019) Magnetic resonance imaging-targeted biopsy versus systematic biopsy in the detection of prostate Cancer: a systematic review and Meta-analysis (figure presented.). Eur Urol 76(3):284–303. https://doi.org/10.1016/j.eururo.2019.04.043
Kwak JT, Hewitt SM (2017) Lumen-based detection of prostate cancer via convolutional neural networks. Med Imaging 2017 Digit Pathol 10140:1014008. https://doi.org/10.1117/12.2253513
Li W, Li J, Sarma KV, Ho KC, Shen S, Knudsen BS, Gertych A, Arnold CW (2018) Path R-CNN for prostate cancer diagnosis and Gleason grading of histological images. IEEE Trans Med Imaging 38(4):945–954
Litjens G, Debats O, Barentsz J, Karssemeijer N, Huisman H (2014) Computer-aided detection of prostate cancer in MRI. IEEE Trans Med Imaging 33(5):1083–1092. https://doi.org/10.1109/TMI.2014.2303821
Litjens G, Debats O, Barentsz J, Karssemeijer N, Huisman H (2017) “ProstateX Challenge data”, Cancer Imaging Archive. https://doi.org/10.7937/K9TCIA.2017.MURS5CL
Moradi M, Abolmaesumi P, Siemens DR, Sauerbrei EE, Boag AH, Mousavi P (2009) Augmenting detection of prostate cancer in transrectal ultrasound images using SVM and RF time series. IEEE Trans Biomed Eng 56(9):2214–2224. https://doi.org/10.1109/TBME.2008.2009766
Schalk SG, Demi L, Bouhouch N, Kuenen MP, Postema AW, de la Rosette JJ, Wijkstra H, Tjalkens TJ, Mischi M (2017) Contrast-enhanced ultrasound angiogenesis imaging by mutual information analysis for prostate Cancer localization. IEEE Trans Biomed Eng 64(3):661–670. https://doi.org/10.1109/TBME.2016.2571624
Singh SK, Goyal A (2020) Three stage cervical cancer classifier based on hybrid ensemble learning with modified binary PSO using pretrained neural networks. Imaging Sci J 68(1):41–55. https://doi.org/10.1080/13682199.2020.1734306
Singh AK, Goyal A (2020) “Performance analysis of machine learning algorithms for cervical cancer detection,” Int J Healthcare Info Syst Info (IJHISI), IGI Global 15(2):1–21
Yan K, Li C, Wang X, Li A, Yuan Y, Feng D, Khadra M, Kim J (2016) “Automatic prostate segmentation on MR images withdeep network and graph model. In Engineering in Medicine and BiologySociety (EMBC)”, IEEE 38th AnnualInternational Conference of the of theIEEE Engineering in Medicine and Biology Society, pp. 635–638
Yang M, Li X, Turkbey B, Choyke PL, Yan P (2013) Prostate segmentation in MR images using discriminant boundary features. IEEE Trans Biomed Eng 60(2):479–488. https://doi.org/10.1109/TBME.2012.2228644
Zeiler MD, Fergus R (2014) “Visualizing and understanding convolutional networks”, In European conference on computer vision pp.818–833
Zeiler MD, Krishnan D, Taylor GW, Fergus R (2010) “Deconvolutional networks,” 2010 IEEE Computer Society Conference on Computer Vision and Pattern Recognition, San Francisco, CA, USA, pp. 2528–2535. https://doi.org/10.1109/CVPR.2010.5539957
Zhang G, Wang W, Yang D, Luo J, He P, Wang Y, Luo Y, Zhao B, Lu J (2019) A bi-attention adversarial network for prostate Cancer segmentation. IEEE Access 7:131448–131458. https://doi.org/10.1109/ACCESS.2019.2939389
Acknowledgments
We gratefully thank AAPM, NCI, SPIE and the Radboud University for giving the prostate-2 dataset in its challenge.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author has declared that there is no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Singh, S.K., Sinha, A., Singh, H. et al. A novel deep learning-based technique for detecting prostate cancer in MRI images. Multimed Tools Appl 83, 14173–14187 (2024). https://doi.org/10.1007/s11042-023-15793-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-15793-0