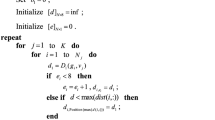Abstract
Superpixel become increasingly popular in image segmentation field as it greatly helps image segmentation techniques to segment the region of interest accurately in noisy environment and also reduces the computation effort to a great extent. However, selection of proper superpixel generation techniques and superpixel image segmentation techniques play a very crucial role in the domain of different kinds of image segmentation. Clustering is a well-accepted image segmentation technique and proved their effective performance over various image segmentation field. Therefore, this study presents an up-to-date survey on the employment of superpixel image in combined with clustering techniques for the various image segmentation. The contribution of the survey has four parts namely (i) overview of superpixel image generation techniques, (ii) clustering techniques especially efficient partitional clustering techniques, their issues and overcoming strategies, (iii) Review of superpixel combined with clustering strategies exist in literature for various image segmentation, (iv) lastly, the comparative study among superpixel combined with partitional clustering techniques has been performed over oral pathology and leaf images to find out the efficacy of the combination of superpixel and partitional clustering approaches. Our evaluations and observation provide in-depth understanding of several superpixel generation strategies and how they apply to the partitional clustering method.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Digital image processing is in high demand in a variety of fields, including computer vision [22], robotics [115], remote sensing [168], industrial inspection [51], and medical diagnostics [129]. Superpixels generation is a technique that is commonly employed as a pre-processing step in computer vision applications. Introduced by Ren et al. [122], superpixels are small image regions with a uniform appearance that are created when the original image is over-segmented. Superpixels can be define as groups of pixels with identical characteristics that can be utilised as mid-level units to reduce computing costs of many computer vision problems, such as image segmentation, saliency, tracking, classification, object detection, motion estimation, reconstruction. The superpixel representation considerably decreases the amount of image primitives and hence enhances the representational efficiency when compared to the regular pixel representation in images. In contrast to pixel, superpixel can greatly reduce the size of the object to be processed as well as the complexity of subsequent processing [3, 149, 150]. Because of these advantages superpixel approaches are commonly employed as a pre-processing step for a variety of tasks [3, 84, 149, 150].
Good superpixels techniques can increase the performance of computer vision applications [145]. There are several methods for producing superpixels, each with its own set of benefits and downsides that may be better suited to a certain application. If the speed of the algorithm is critical, the SEEDS [138] algorithm may be a better option. If superpixels are utilised to form a graph, the Normalized Cuts (NC) [126] algorithm may be the best option, as it generates more regular superpixels with a more attractive appearance. If superpixels are utilised as a pre-processing step in segmentation algorithms, a method that considerably improves segmentation algorithm performance, such as SLIC [5], is likely a viable choice. While defining an optimal method for all applications is challenging, the following characteristics are generally desirable: (a) Superpixel boundaries should correspond strongly to object boundaries, allowing as many pixels on the object boundaries to be recalled as feasible. (b) Superpixels should be quick to compute, memory efficient, and simple to use when employed to minimise computational complexity as a pre-processing step. (c) Superpixels should improve both the speed and the quality of results when utilized for segmentation purpose.
Image segmentation can be accomplished by a variety of clustering algorithms. However, there is no agreed-upon definition of a cluster, and the many methods of clustering have each developed their own strategies for grouping the data. Therefore, the clustering techniques can be roughly divided into two groups depending on the cluster formation: hierarchical clustering and partitional clustering. Figure 4 displays a taxonomy of the clustering techniques. Because of its superior computational efficiency, partitional clustering has gained a significant amount of popularity in recent years and is now widely considered to be superior to hierarchical clustering. This is especially true for larger datasets. In this paper we use clustering algorithms, specially the partitional clustering, for image segmentation. The selected partitional clustering algorithms are: the K-means, the Fuzzy c-Means (FCM).
The paper provided here is much more focused on the fundamentals; it is based on a comparison of several clustering techniques, both with and without superpixel pre-processing, using the same dataset containing 100 different Rosette Plant images and 100 different Oral histopathology images. As SLIC algorithm is mostly used among others superpixel generation technique for image pre-processing, we use this algorithm for superpixel generation. The images are segmented using K-Means (KM), Fuzzy C-Means (FCM), Genetic Algorithm (GA) and Particle Swarm Optimization (PSO) Algorithms. We also compare the above methods with Multiscale Superpixel based Fast Fuzzy C-Means Clustering (SFFCM) method. The images are segmented and their binary forms are compared to the Ground Truth Image, with the determining factor being the average value of all the algorithms.
Research questions
-
1)
In a wide variety of computer vision applications, superpixel segmentation proved to be an effective step in the pre-processing task. The goal of superpixel is to decrease image redundancy and boost efficiency from the perspective of the next processing task. There are numerous methods for creating superpixels, each with advantages and disadvantages that may be better suited for a specific purpose. So, choosing the right superpixel algorithm for a particular application is a major research topic. In superpixel based segmentation the following characteristics are often favoured: (i) The boundaries of an image should be well-adhered by superpixels. (ii) Superpixels should be fast, memory-efficient, and easy to utilise while reducing computational complexity.
-
2)
Which specific superpixel generation technique has to be chosen for a specific image type?
-
3)
Which superpixel generation algorithm is computationally effective?
-
4)
How to identify what no of superpixel is best for a specific image type?
-
5)
There are mainly two types of clustering techniques: Partitional clustering and Hierarchical clustering. Partitional clustering has grown significantly in popularity in recent years and is currently seen as being superior to hierarchical clustering due to its higher processing efficiency. But it has three major drawbacks, i) Higher computational time ii) Local optima trapping and iii) Sensitive to noise. Therefore, finding the strategies to overcome the said drawback will be an important task.
-
6)
How the noise robustness can be incorporated into the partitional clustering algorithms?
-
7)
How to overcome the local optima trapping problem of partitional clustering algorithms?
-
8)
How the computational efficiency can be improved for partitional clustering algorithms?
-
9)
In the literature, it is noticed that the use of superpixel as a pre-processing task greatly reduces the computational time. It also performs well for noisy image segmentation. So, the incorporation of superpixel image as the input of the partitional clustering algorithms might overcome the problems of higher computational time and sensitivity to noise. But finding the optimal combination of superpixel generation techniques and partitional clustering algorithms is an important task.
-
10)
Which combination of superpixel generation techniques and partitional clustering algorithms is optimal for image segmentation?
The paper is organized as follows: Section 2 presents the literature survey and datasets part. Section 3 demonstrates the superpixel generation technique, Section 4 represents clustering methodologies. Section 5 represents the experimental results. The paper is concluded in Section 6.
2 Literature survey and datasets
This segment provides the current image segmentation and various superpixel generation techniques with their corresponding datasets, as shown in Tables 1 and 2, respectively. Superpixel based image clustering is a method of partition an image into multiple segments. The objective of superpixel based image clustering is to represent image something more constructive and simpler to examine. So, a detailed survey on various papers is necessary to check the need for a new framework. Literature survey made on various papers which includes the techniques of image segmentation like KM, FCM, GA, PSO etc. and various superpixel generation algorithm like SLIC, MMGR-WT, Ncuts etc.
2.1 Paper sources and keywords
-
A)
Sources
The mentioned papers have been collected from the following sources:
-
(i)
Google Scholar—https://scholar.google.com
-
(ii)
IEEE Xplore—https://ieeexplore.ieee.org
-
(iii)
ScienceDirect—https://www.sciencedirect.com
-
(iv)
SpringerLink—https://www.springerlink.com
-
(v)
ACM Digital Library—https://dl.acm.org
- (vi)
-
B)
Keywords
Each of these above sources is queried with the following combinations of keywords:
-
KW1: Superpixel image-based segmentation
-
KW2: Clustering based image segmentation
-
KW3: Hierarchical and Partitional clustering based image segmentation
-
KW4: Nature Inspired Optimization Algorithm based image clustering
-
KW5: Noisy image segmentation using clustering techniques
-
KW6: Image segmentation using superpixel image-based clustering technique
-
KW7: Computationally effective and noise robust image segmentation
-
KW8: Image segmentation using K-Means
-
KW9: Image segmentation using Fuzzy C-Means
-
KW10: Image segmentation using Swarm Intelligence algorithms
The graphical analysis has been conducted based on the application of superpixel image for clustering-based image segmentation. The following are the top three factors that were taken into consideration in the graphical analysis: (a) Year wise development of superpixel image for clustering-based image segmentation techniques which has been shown in Fig. 1. The figure represents the year wise development statistics of last five year which clearly shown that the utilization of superpixel image for clustering-based image segmentation is continuously popular. (b) A category wise percentage of various superpixel generating techniques has been presented in Fig. 2. From Fig. 2, it is clearly noticed that one third of applications have been done over the SLIC superpixel generating technique. MMGR-WT is the next most commonly used superpixel generating technique. (c) The percentages of various clustering approaches that have been used are shown in Fig. 3 which shows that FCM is the most used clustering technique, followed by KM, NIOA, and other techniques.
3 Discussion on superpixel generating techniques
The objective of superpixel segmentation is to partition pixels in an image into atomic areas with bounds that match the natural object boundaries. In computer vision, superpixel segmentation has sparked a lot of attention because it makes it easy to compute picture attributes and reduces the complexity of future image processing tasks. In recent times, a slew of superpixel segmentation methods have been introduced. Superpixel segmentation techniques are classified into numerous groups based on various categorization criteria. They are described below in detail.
3.1 Density-based methods
Density-based algorithms [130] are often classed as over segmentation techniques since they lack control on the amount of superpixels or their compactness. Edge-Augmented Mean Shift (EAMS) [23] and Quick Shift (QS) [139] are two popular density-based methods. In a calculated density image, both execute mode-seeking, every pixel is specified to the method it falls into.
3.2 Watershed-based methods
These methods are based on the watershed algorithm (WT) [98, 130] and typically vary in how the image is pre-processed and how markers are set. The number of superpixels is determined by the number of markers, and some watershed-based superpixel methods provide compressibility control., for example Water Pixel (WP) [95] or Compact Watershed (CW) [108]. Morphological Superpixel Segmentation (MSS) [11] is also a popular density based superpixel algorithm.
3.3 Graph-based methods
Graph-based algorithms [68, 148] consider the image as an undirected graph and divide it into sections based on edge-weights, which are frequently computed as colour differences or similarities. The partitioning techniques differ; for example, Felzenswalb and Huttenlocher (FH) [46], Entropy Rate Superpixels (ERS) [92], and Proposals for Objects from Improved Seeds and Energies (POISE) [67] merge pixels into superpixels from the bottom up, whereas Normalized Cuts (NC) [126] and Constant Intensity Superpixels (CIS) [140] utilise cuts, and Pseudo-Boolean Optimization Superpixels (PB) [162] uses elimination.
3.4 Path-based methods
Path-based methods divide an image into superpixels by connecting seed points to pixel paths that satisfy specific standards. The number of superpixels can be conveniently manipulated, but not their density. These algorithms [43, 130] frequently make use of edge data: Topology Preserving Superpixels (TPS) [48, 133] employs edge detection while Path Finder (PF) [43] uses discrete image gradients.
3.5 Contour evolution-based methods
Contour evolution is one of the popular superpixel generation algorithm [130]. Starting with an initial seed pixel, these techniques portray superpixels as developing outlines. Turbo Pixel (TP) [86] and Eikonal Region Growing Clustering (ERGC) [14] are the example of contour evolution-based method.
3.6 Energy optimization-based methods
These algorithms [130] maximise a formulated energy in a step-by-step manner. As an initial superpixel segmentation step, the image is divided into a regular grid, and based on energy, pixels are interchanged among nearby superpixels. The amount of superpixels can be adjusted, the compactness can be adjusted, and iterations can usually be stopped at any time. Some of the Energy optimization method are Contour Relaxed Superpixels (CRS) [25], Superpixels Extracted via Energy Driven Sampling (SEEDS) [138], Convexity Constrained Superpixels (CCS) [134] and Extended Topology Preserving Segmentation (ETPS) [158].
3.7 Clustering-based methods
These superpixel generation technique [130] are based on clustering algorithms such as k-means, which use colour, spatial, and extra information such as depth and are initialised by seed pixels. The amount of created superpixels and their compactness are intuitively configurable. Although these techniques are iterative, connection must be enforced through post-processing. Some of the clustering-based method is Depth-Adaptive Superpixels (DASP) [146], Vcells (VC) [142] and Simple Linear Iterative Clustering (SLIC) [4, 5]. DASP uses extra depth information. SLIC algorithm generates superpixels by clustering pixels based on their colour similarity and proximity in the image plane. It is one of the most often used algorithms for superpixel generation. SLIC is time efficient. Using a kernel function, Linear Spectral Clustering (LSC) [87] converts image pixels into weighted points in 10-D feature space. The seed pixels are then uniformly sampled for the entire image. The search centres are then used, and the feature vectors are used as the initial weighted means of the relevant clusters. LSC use the spectral clustering technique to arrive at a global optimal solution, which improves boundary adherence even more. Pre-emptive Simple Linear Iterative Clustering (PreSLIC) [114] is also a popular clustering-based superpixel generation algorithm which is proposed by Neubert et al. [109]. To prevent revisiting clusters, Pre-emptive SLIC implements a local terminal criterion for each cluster. As a result, clusters are only updated if there are significant changes in the cluster centre.
4 Clustering techniques based image segmentation
Clustering is a powerful image segmentation technique that has been developed. The taxonomy of the clustering techniques is displayed in Fig. 4. The goal of cluster analysis is to divide an image data set into a number of disjoint groups or clusters with greater intra-cluster similarity but lower inter-cluster similarity. It is obvious that intra-cluster similarity should be enhanced while inter-cluster similarity should be reduced. Based on this concept, objective functions are formulated [30, 36, 39]. The method of minimizing/maximizing one or many objective functions can be used to achieve the better partitioning of a provided data collection. The objective functions are generated by establishing a statistical–mathematical relationship between the independent data items and the proposed set of cluster representatives [30, 36, 39]. The partitions into c classes should preserve the respective characteristics:
-
1.
At least a single vector should be assigned to each cluster. i.e.,
$$ {c}_i\ne \varnothing, \forall i\in \left\{1,2,\dots, c\right\} $$ -
2.
There should be no data vector in common between two distinct clusters. i.e.,
$$ {c}_i\cap {c}_j=\varnothing, \forall i\ne j\ \mathrm{and}\ i,j\in \left\{1,2,\dots, c\right\} $$ -
3.
Each data vector should absolutely be associated with a cluster. i.e.
$$ {\cup}_{i=1}^c={D}_v $$
Where, Dv is the total number of objects.
Clustering-based image segmentation techniques are classified [124]
The clustering techniques are classified into following categories:
4.1 Hierarchical clustering
In this clustering method, data is grouped at multiple levels of comparability, which is represented as a dendrogram, a tree-like structure in Fig. 5. In general, there are two techniques to hierarchical clustering: divisive and agglomerative [103, 124]. Hierarchical clustering can be accomplished in two ways. They can be bottom-up or top-down. Large clusters are divided into small clusters, and small clusters of large clusters are combined together. The methods in these two sub-categories are summarised in Table 3. The pros and cons of the hierarchical clustering are as follows:
Categories of hierarchical clustering [103]
The Pros are:
-
The algorithm does not require the number of clusters (k) to be pre-defined.
-
Integrated adaptability with respect to the degree of granularity.
-
Creates more comprehensible clusters, which could be beneficial to the discovery.
-
Extremely well suited with regard to issues concerning point linkages.
-
Allows for the use of any standard units of measurement.
-
Beneficial for the presentation of data, as it provides a hierarchical relation between clusters.
The Cons are:
-
When used to massive amounts of data, hierarchical clustering is not very effective.
-
The algorithm is sensitivity to noise and outliers.
-
Once a decision is made in hierarchical clustering, it cannot be reversed.
-
When dealing with varying cluster sizes, the algorithm encounters difficulty.
-
When the decision to split or merge is made, there is no way to go back and change it.
-
The sequence of the data has an impact on the final outcomes with this technique.
-
A time complexity of at least O(n2 log n) is required, where n is the number of data points.
4.1.1 Divisive clustering
Divisive clustering is a top-down strategy that begins with a singletons cluster or model including all data points and recursively divides it. Initially, all of the data items were assigned to a singletons cluster. This singletons cluster is subdivided into small cluster still a stopping criterion is met or each data item becomes its own cluster. This clustering technique is more complicated than agglomerative clustering because it splits the data till each cluster has only one data item. If every cluster is not divided into distinct data leaves, divisive clustering method will be computationally proficient. This has high computing costs and model selection difficulties, just like agglomerative clustering. Furthermore, because data might be divided into two clusters in the first phase, it is quite sensitive to initialization. Divisive clustering has a high temporal complexity (n2). Furthermore, divisive clustering is more accurate than top-level partitioning since it considers the global distribution of data while partitioning data. Popular approaches in this area include divisive clustering (DIVCLUS-T) [18], Divisive information theoretic feature clustering (DITFC) [42] and divisive hierarchical clustering with diameter criterion (DHCDC) [62], Cell-dividing hierarchical clustering (CDHC) [90], Stochastic Multi-objective Acceptability Analysis (SMAA) - Multicriteria Divisive Hierarchical Clustering (SMAA-MDHC) [71] and Variance-Cut (VC) [16]. The pros and cons of the divisive clustering are as follows:
The pros are:
-
Additionally, it is not necessary to provide a number of clusters in advance while using this approach.
-
The divisive algorithm gives more accurate result by taking global distribution of data when making top-level partitioning decisions.
-
When making top-level partitioning decisions, divisive clustering considers the global distribution of data.
-
Divisive clustering is more efficient if we don’t create a complete data hierarchy.
The cons are:
-
Rigid, i.e., once a splitting operation has been performed on a cluster, it cannot be undone.
-
There is no prior information concerning the minimum required number of clusters.
-
Because of the large space and time complexities, the techniques are not suitable for huge datasets.
-
The algorithm is Sensitivity to noise and outliers.
4.1.2 Agglomerative clustering
This clustering technique follows bottom-up strategy in which each entity represents its own cluster, which is subsequently combined iteratively until the desired cluster structure is attained. The N-sample algorithm begins with N clusters, each of which contains a single sample. Then, until the number of clusters is decreased to one or the user specifies, two clusters with the greatest similarity will be combined. The parameters used in this approach are minimum, maximum, average, and centre distances. A naive agglomerative clustering has a time complexity of O(n3), which can be decreased to O(n2) utilising optimization procedures. Single Linkage (SLINK) [60], Complete Linkage (CLINK) [125], Average Linkage or Unweighted Pair Group Algorithm with Arithmetic Mean (UPGMA) [61], Weighted Pair Group Algorithm with Arithmetic Mean (WPGMA) [105], Unweighted Pair Group Method Centroid (UPGMC) [13], Balanced iterative reducing and clustering using hierarchies (BIRCH) [161], clustering using representatives (CURE) [63], and CHAMELEON [75] are three common agglomerative strategies. The pros and cons of the agglomerative clustering are as follows:
The Pros are:
-
There is no need to pre-specify the number of clusters in agglomerative clustering.
-
It uses its decisions on the differences that exist between the things that are to be classified together.
-
It can generate an object ordering that may be useful for the display.
-
The Agglomerative Clustering method will break the data up into smaller clusters, which may discover similarities in data.
The Cons are:
-
Only in certain circumstances does the agglomerative approach produce the greatest results.
-
The method can’t undo previous work, so if items were erroneously sorted, the same outcome should be nearby.
-
Varied distance metrics can provide different results when calculating distances between clusters.
-
Due to the high time and space complexity, it is not suitable for huge datasets.
Algorithms 1 and 2 show the pseudocode for the divisive and agglomerative clustering techniques, respectively
4.2 Partitional clustering techniques
Due to its computing efficiency, partitional clustering is more common and preferable than hierarchical clustering, particularly for huge datasets. In this clustering approach, the idea of commonality is used as the measuring parameter. In general, partitional clustering divides data into clusters based on an objective function, with data from one cluster being more equivalent to data from other clusters. To accomplish this, the commonality of every data item to each cluster is calculated.
Furthermore, in partitional clustering, the overall concept of the objective function is the minimization of the within-cluster commonality factors, which is commonly calculated utilising Euclidean distance. The objective function represents the quality of each formed cluster and produces the finest presentation from the clusters that have been generated. Furthermore, partitional clustering algorithms ensure that every data element is allotted to a cluster, even if it is located much further from the cluster center. This can lead to cluster shape distortion or erroneous results, especially when there is noise or an outlier.
Image segmentation, robotics, wireless sensor networks, web mining, corporate management, and medical sciences are just a few of the fields where partitional approaches have been used. The data distribution and complexity vary by application domain.
As a result, a single partitional clustering method may not be appropriate for all cases. As a result, the appropriate method is chosen based on the problem and dataset. Hard clustering and soft clustering are the two basic types of partitional image clustering techniques. A pixel becomes a part of a single cluster in hard clustering. As a result, there is a binary membership (i.e., 0 or 1) between clusters and elements. This technique calculates all of the cluster centres and then assigns the elements to the closest cluster. One of the most notable examples of hard clustering is K-means [113]. Soft clustering, on the other side, uses fractional membership, as opposed to hard clustering, which makes it more practical for real-world applications. A pixel can belong to multiple clusters with varying degrees of membership, which are denoted by fractional membership. Fuzzy C-means (FCM) is one of the examples of soft clustering technique that was suggested by Bezdek [12]. The FCM outperforms hard clustering techniques such as K-means in terms of its ability to deal with ambiguity in grey levels.
4.2.1 Classical K-means (KM) Clustering: A pixel based image segmentation strategy
Suppose, we want to segment an image I consisting of N number pixels and L number of gray levels into K clusters. The image I can be regarded as data point set of pixels. Let, zp indicates the d components of pixel p. Where d indicates the number of spectral bands presents in the image I. For gray and RGB color image d = 1 and d = 3 respectively.
The K-means algorithm (KM) is a popular clustering technique used for image segmentation in computer vision. KM minimizes the following criteria or objective function [27]:
Where, ‖.‖ is an inner product-induced norm in d dimensions which measures the distance among ith pixel \( {\mathbf{z}}_p^i \) and jth cluster center mj. Membership uij = 1 for pixel \( {\mathbf{z}}_p^i \) if it belongs to cluster Cj; otherwise, uij = 0.Now, it is a minimization problem with two steps. At first, we should derogate the objective function J w.r.t. uij by considering the fixed mj. Then the J is minimized w.r.t. mj by considering the fixed uij.
Therefore, the two steps are as follows:
Therefore, we need to assign the pixels to the closest cluster where the closeness is measured by the span between the concerned pixel and cluster centers. The second step is
So, recalculation of cluster centers is necessary to divulge the new pixel assignments to the clusters. Relying on the discussion, the algorithm of K-Means clustering is presented as in Algorithm 3.
4.2.2 Classical fuzzy C-means (FCM) clustering: A pixel based image segmentation strategy
Suppose, we want to segment an image I consisting of N number pixels and L number of gray levels into K clusters. The image I can be regarded as data point set of pixels. Let, zp indicates the d components of pixel p. Where d indicates the number of spectral bands presents in the image I. For gray and RGB color image d = 1 and d = 3 respectively.
The fuzzy C-means algorithm (FCM) [26, 39, 111] utilizes the principles of fuzzy sets to generate a partition matrix or membership matrix(U) while minimizing the following measure or objective function:
Where, ‖.‖ is an inner product-induced norm in d dimensions which measures the distance among ith pixel \( {\mathbf{z}}_p^i \) and jth cluster center mj. Membership or partition matrix U = [uij]N × K and \( {\sum}_{j=1}^K{u}_{ij}=1 \). Fuzzy exponent parameter e and 1 ≤ e ≤ ∞. FCM initializes the cluster centers randomly, and then at each iteration, it computes the fuzzy membership of each pixel using the following expression:
The cluster centers are calculated by using the expression given below:
Depending on the discussion, the algorithm of FCM clustering is presented as Algorithm 4.
4.2.3 Three global challenges of KM and FCM
Although KM and FCM are common clustering techniques, they have three significant shortcomings, which are as follows:
-
1.
Higher Computational Time: In the clustering procedure, the distance among all N pixels of the image and the K cluster centres is calculated repeatedly. As a result, as the image size and number of clusters grow larger, a large amount of computational effort is required. Generally, the time complexity of the KM [27, 113] and FCM [39, 85] is O(N × K × d × t). Where, N, d, and Kdenotes the number of pixels, clusters, and dimension correspondingly and generally, d, K ≪ N. As a result, image size is a major consideration. Finally, t denotes the number of iterations required to finish the clustering process.
-
2.
Local optima trapping: Because of the random centre initialization, KM and FCM are avaricious in nature and commonly focuses to local optima, resulting sin vacuous clusters. Because it is a local optimizer, its efficiency is heavily influenced by the original cluster center selection. Various initializations might result in dissimilar clusters. As a result, KM’s and FCM’s convergence into global optima is a major problem. The numbers of ways of N pixels are sundered into K non-empty clusters are represented by Stirling numbers of the second type:
$$ S\left(N,K\right)=\frac{1}{K!}\sum \limits_{i=0}^K{\left(-1\right)}^{K-i}\left(\genfrac{}{}{0pt}{}{K}{i}\right){i}^N $$(7)
It can be easily noticed that the number of ways of partition can be approximated by \( \frac{K^N}{K!} \) which is colossal number. As a result, the entire enumeration of every potential clustering to identify the global optima of Eq. (1) is clearly computationally prohibitively expensive, unless for very smaller image. In addition to that, it is proved that this non-convex minimization problem is NP-hard even for d = 2 or K = 2.
FCM is greedy by nature, and because to the random centre initialization, it commonly accumulates to local optima and might even yield empty clusters [26]. Because it is a local optimizer, its efficiency is heavily influenced by the initial cluster center choosing. Various initializations might result in dissimilar clusters. As a result, in FCM, convergence into global optima is a severe issue. It is easy to demonstrate like in K-means that the full enumeration of every potential clustering to discover the global optima of Eq. (4) is unquestionably computationally too expensive, and this non-convex minimization problem is NP-hard.
-
3.
Sensitive to Noise: The distributed features of the pixels have a major impact on the convergence rate of KM and FCM. If the image’s histogram is identical, it’s challenging to discover ideal cluster centres in a short amount of time [27, 39, 85]. However, A histogram with multiple noticeable peaks is simple to cluster. Furthermore, KM and FCM produce poor results for noisy images and take a long time.
4.3 Problem solving strategies
Researchers analyse the three major problems with KM and FCM and report some solutions in the literature. The following section discusses the reported solution strategies for the aforementioned problems.
4.3.1 Image histogram based clustering: A time reduced approach
Both KM and FCM are associated with a good deal of time complexity and this is the main reason for the degradation of its performance with the very large dataset. It is previously mentioned that the time complexity of KM and FCM is O(N × c × d × t), where, N, d, and Kdenotes the number of pixels, clusters, and dimension correspondingly, and generally, d, K ≪ N. As a result, the size of the image is an enormous matter. The number of iterations required to finish the entire grouping process is denoted by t. To avoid the time complexity problem, segmentation was performed on a grey level histogram rather than image pixels in this paper. Because the grey levels are frequently much less than the number of pixels in the image, the computation time is short. As a result, the histogram-based KM and FCM are presented as follows:
-
(a)
Histogram based K-Means (HBKM) Algorithm: A Time Reduced Hard Clustering Strategy
To overcome the significant time complexity problem, this Histogram based K-Means (HBKM) approach clusters on grey level histograms rather than pixels in the image. As a result, the computing time is short because the number of grey levels in an image is typically much fewer than the number of pixels [27, 40]. As a consequence, the objective function may be written as follows:
Where, L indicates the number of grey levels of the image. As an example, there are 256 different grey levels in an 8-bit image. ‖.‖ is an inner product-induced norm in d dimensions which compute the distance between gray level gi and cluster center mj and γi is the total number of pixels with gi gray level and hence, \( \sum \limits_{i=1}^L{\gamma}_i=N \) and 1 ≤ i ≤ L. Membership uij = 1 for pixel \( {\mathbf{z}}_p^i \) if it belongs to cluster Cj; otherwise, uij = 0. Mathematically, in the same way the memberships of the gray levels are calculated as follows:
Therefore, we have to disperse the grey levels to the adjoining cluster where the closeness is measured by the distance between the concerned grey levels and cluster centers. These cluster centers are computed by the following expressions:
So, revaluations of cluster centers are obligatory to reveal the new gray level assignments to the clusters. Based on the above discussion, the algorithm of histogram-based K-Means (HBKM) clustering is presented as in Algorithm 5.
-
(b)
Histogram based Fuzzy C-Means (HBFCM) Algorithm: A Time Reduced Soft Clustering Strategy
To overcome the huge time complexity issue, this Histogram based Fuzzy C-Means (HBFCM) [39, 40, 85] method clusters grey level histograms rather than pixels in the image. As a result, because grey levels are typically significantly less than the number of pixels in an image, the computing time is low. Hence, the objective function can be described as
Where, L denotes the number of gray levels of the image. As an example, there are 256 different grey levels in an 8-bit image. ‖.‖ is an inner product-induced norm in d dimensions which compute the distance between gray level gi and cluster center mj and γi is the total number of pixels with gi gray level and hence, \( \sum \limits_{i=1}^L{\gamma}_i=N \) and 1 ≤ i ≤ L. Fuzzy exponent e and 1 ≤ e ≤ ∞. The members. Membership or partition matrix U = [uij]L × K and \( {\sum}_{j=1}^K{u}_{ij}=1 \). The following is how the membership and centres are calculated:
Because of histogram-based clustering, a membership partition matrix U = [uij]L × K is obtained. But, uij is a fuzzy membership of gray value i with respect to cluster j, a new membership partition matrix \( {\mathrm{U}}^{\prime }={\left[{\mathrm{u}}_{\mathrm{ij}}^{\prime}\right]}^{\mathrm{N}\times \mathrm{K}} \) which corresponds to the original full image I(x, y), is obtained by:
where, I(x, y) indicates intensity value of original image I at location (x, y).
As a result, all the pixels with intensity value gi correlated with membership value \( {u}_{ij}^{\prime } \). Based on the analysis, the algorithm for histogram based fuzzy C-Means (HBFCM) clustering is described as Algorithm 6.
In the case of HBKM and HBFCM, some pros and cons which are described below.
-
1.
Time Complexity: The key benefit is that reduce the time complexity. The time complexity of HBKM and HBFCM is O(L × K × d × t), and L ≪ N in an image. Where L is determined by the number of bits assigned to each pixel. For an 8-bit image, the value of L is always 256. As a result, the clustering method is independent of image size, resulting in a significant reduction in time complexity when using this histogram-based image clustering method. On the other hand, HBFCM and HBKM have two major drawbacks, one is sensitive to noise and the other is local optima trapping.
-
2.
Local Optima Trapping: Because of the random center initialization, HBKM and HBFCM are greedy in nature and converge to local optima frequently. The initial cluster center selection has a significant impact on the efficiency of the HBKM and HBFCM. Different clusters may result from different initializations. As a result, convergence into global optima is a severe issue in both HBKM and HBFCM. In HBKM, the number of ways of L grey levels are partitioned into K non-empty clusters which is represented by Stirling numbers of the second form:
It can be easily seen that the number of ways of partition can be approximated by \( \frac{K^L}{K!} \) which is also a big number although L ≪ N.As a result, finding the global optima of Eq. (8) using the whole inventory of every conceivable clustering is computationally expensive. As a result, this non-convex minimization issue qualifies as NP-hard.
It is true for HBFCM that although L ≪ N, however, the entire enumeration of all potential clustering to obtain the global optima of Eq. (11) is unquestionably computationally costly. As a result, this non-convex minimization issue qualifies as NP-hard.
-
3.
Sensitive to Noise: It is previously stated that A histogram with multiple noticeable peaks is straightforward to cluster. However, noise obliterates the histogram’s peaks and alters the features of the pixel’s dispersal throughout the image. As a result, clustering based only on histograms produces poor results and takes a long time.
-
4.
Problem with Color image: Extending the histogram-based clustering technique to segment color images is difficult due to the complexity of determining the color image’s histogram [112]. Another reason is that the number of different colors is large and hence, it often goes near to the number of pixels present in a color image. Superpixel can be utilized to deliberate this problem.
4.3.2 Noise robust K-means and fuzzy C-means approaches
The summaries of strategies that are utilized to make KM and FCM noise-robust are as follows: (i) incorporation of spatial information, or adaptive spatial information, or weighted spatial information into KM and FCM; (ii) filtering of noisy images before the commencement of actual clustering process. Some notable filtering techniques are Mean filter, median filter, non-local mean filter, morphological reconstruction, bilateral filter; (iii) Filtering of membership [39, 85] or incorporation of spatial information into membership function of FCM [93].
For example, Ahmed et al. [6] proposed the FCM method considering spatial constraints (FCM_S) based on local spatial information. In FCM_S, the cost function of the original FCM has been modified by considering the inhomogeneity intensity and the labelling of a pixel influenced based on the labels in its nearest immediate neighbourhood. The main issue of FCM_S was its large computational time due to the computation of local information median filter. However, both the variant unable to deliver satisfactory outcomes over different noise-corrupted images. Szilagyi et al. [35] developed an Enhanced FCM (EnFCM). EnFCM is based on the linearly weighted sum image, which is estimated from the input image and the average gray level of the local neighbourhood of each pixel. Here, the clustering is performed using the gray level histogram. As a result, a reduced computational time is observed. However, the segmentation accuracy of EnFCM is only comparable to FCM_S. The segmentation accuracy of EnFCM depends on the parameter α (or λ), the size of the chosen window and the filtering method used. Krinidis and Chatzis [82] proposed a parameter-free FCM variant called fuzzy local information c-means clustering algorithm (FLICM), which incorporated a state-of-art fuzzy factor into the cost function to deliberate the parameter α (or λ) of EnFCM. The fuzzy factor enhanced the ability of noise reduction and sharpness preservation. The main demerit of FLICM was the utilization of fixed spatial distance, which degraded the robustness. Gong et al. [58] incorporated a local variable coefficient instead of static to develop a superior variant of FLICM called Reformulated FLICM (RFLICM). Later, the same author developed another variant of FLICM (KWFLICM) [59] by incorporating a trade-off mechanism of weighted fuzzy factor and a kernel metric to increase the performance regarding noise reduction and outliers. But KWFLICM was a parameter-free variant but had a higher computational time compared to FLICM.
The approaches mentioned above suffer from high computational costs due to the repetitive computation and incorporation of local spatial information into objective function. In order to reduce the cost of computation without discarding local spatial information, proposed by Chen and Zhang [19] two variants of FCM_S known as; FCM_S1 and FCM_S2 depending on the use of mean and median filters respectively over the image before the starting of iteration stage. Therefore, time was reduced because of the pre-computation of local spatial information of the image. As FCM_S1 utilized a mean filter, it was robust to Gaussian noise. On the other hand, FCM_S2 robust to salt & pepper noise due to the utilization of the median filter. A fast and robust FCM (FRFCM) in [85] proposed by Lei et al., where a morphological reconstruction-based (MR) approach was applied to reduce the error due to noise. Niharika et al. [110] utilized median filter before applying K-means based clustering in Synthetic Aperture Radar (SAR) image segmentation field. Due to the application of median filter, K-means utilized not only the gray level information, but spatial information also. Finally, morphological closing reconstruction was incorporated to provide accurate segmentation. The experimental results proved that the proposed segmentation technique associate with less error percentage in a noisy environment. In addition to that, some improved or adaptive K-means variants had been developed. For example, Yao et al. [157] proposed an improved K-means algorithm by combining the traditional K-means, Otsu technique, and morphological reconstruction for the accurate segmentation of fish images. At first, the number of clusters was determined by the number of prominent gray histogram peaks. Then cluster centers computed by traditional K-means were filtered by comparing the mean with the threshold computed by Otsu. Finally, morphological opening and closing operations had been employed to find the contour of the fish body.
Filtering of membership also makes the FCM noise robust. For instance, median filtering has been applied to the membership matrix to decrease pixel misclassification due to noise. The membership filtering can be done on each iteration, but this will significantly increase the processing time, making the method more complex. As a result, membership filtering is performed only once over the final membership matrix generated, resulting in a reduction in computing effort. In reality, the fuzzy C-means use local spatial information in the same way as membership filtering does to improve segmentation accuracy. Membership filtering [26, 111] can be used to take the place of incorporating local spatial information.
4.3.3 Superpixel image based segmentation: A noise robust and time reduced approach
Although many updated algorithms solve the issue by including local spatial information in the cost function, this promotes higher computational complexity. The above said FCM variants are challenging to apply for color images. Fortunately, superpixel [92] can handle the problem. Superpixel is a pre-processing image task which over-segments an image into several smaller sections. In an image, a superpixel region is typically characterised as homogenous and cognitively uniform sections [143]. Due to two benefits, Superpixel can improve image segmentation effectiveness and efficiency. On the one hand, superpixel can do image pre-segmentation based on the images’ local spatial information. Commonly, neighbouring windows used by FCM S, FLICM, FGFCM, KWFLICM, NWFCM, NDFCM, and FRFCM provide poorer local spatial information than the pre-segmentation obtained by superpixel. Superpixel, on the other hand, can reduce the number of different pixels in an image by replacing all pixels in a region with the superpixel region’s mean value [20, 80]. Some basics superpixel generators can be found in [130]. Lei et al. [84] developed a superpixel-based fast FCM clustering technique (SFFCM) for color image segmentation that was substantially faster and more robust than existing clustering techniques. To create a superpixel image with correct contour, a multi-scale morphological gradient reconstruction (MMGR) procedure was initially performed. In contrast to traditional adjacent windows of fixed shape and size, the superpixel image delivers more adaptive and irregular local neighbours that help in color image segmentation. Second, the original color image is effectively condensed based on the generated superpixel image, and its histogram is easily generated by counting the number of pixels in each superpixel region. Ultimately, the superpixel image was subjected to FCM using the histogram parameter to achieve the final segmentation result. The experimental results over the noisy synthetic image and general color images demonstrated that the SFFCM was superior to state-of-the-art clustering algorithms. Wu et al. [149] presented an improved SFFCM approach (ISFFCM), which substitutes fuzzy simple linear iterative clustering for the MMGR in SFFCM (Fuzzy SLIC). For the majority of types of noise, such as Gaussian, multiplicative and salt and pepper, Fuzzy SLIC outperforms MMGR in terms of performance and robustness. Anter and Hassenian [8] employed FCM over adaptive watershed generated superpixel images. The findings reveal that the proposed FCM variant takes less time to compute and performs better on non-uniform CT images by considering less sensitivity to noise. Kumar et al. [83] have suggested a super-pixel-based FCM (SPOFCM) strategy that incorporates the consequences of spatially neighbouring and equivalent superpixels and yielded excellent results.
Another usefulness of superpixel is the reduction of time complexity of FCM when segmenting color as well as gray images. Histogram based clustering is tough to apply for color images. The number of regions in the super-pixel image is significantly less than the original color image’s pixel count. Therefore, the computational time can be reduced to some extent by using superpixel images especially for color images.
-
(a)
Clustering based Superpixel image Segmentation
Clustering based on superpixels for color image segmentation [84], the objective function is taken as:
Where, l is color level and 1 ≤ l ≤ ns, the number of regions in the superpixel image is given by ns. Sl is the number of pixels in the lth region Rl, and zp is the color pixel within the lth region of the superpixel acquired by any superpixel generating method.
For hard clustering like K-Means, the membership U and centers have been computed as follows:
For soft clustering like FCM, the membership U and centers have been computed as follows:
The K cluster center mk has been computed by the Eq. (17). Therefore, the algorithms for superpixel based KM, FCM, and NIOA are represented as Algorithm 7, 8, and 9 respectively.
4.3.4 Nature-Inspired Optimization Algorithms (NIOA) based Clustering: An efficient way to tackle local optima trapping
It is previously noticed that although we are able to time complexity but both classical and histogram-based KM and FCM are regarded as NP-hard and frequently stuck into local optima. The main contribution to solve this local optimum trapping problem is the utilization of Nature-Inspired Optimization Algorithms (NIOA) which are optimization methods used to find the optimal solution for complex problems where mathematical approaches are ineffective [32, 37, 38, 41].
By simulating the behaviour of natural and biological systems, nature-inspired optimization algorithms have been developed. They are simple and effective techniques for dealing with very non-linear and multi-modal real-world issues. According to the No Free Lunch (NFL) theory, just because one algorithm is good at one type of problem does not mean it will be good at all types of problems [147]. As a result, a slew of NIOAs have sprouted up in several optimization fields. Some well-known NIOA are Genetic Algorithm (GA) [57], Differential Evolution (DE) [116], Ant colony optimization (ACO) [96], Particle swarm algorithm (PSO) [44], Artificial bee colony (ABC) [74], Firefly algorithm (FA) [153], Cuckoo Search [156], Bat Algorithm [154], Flower Pollination Algorithm [155] etc. In [our five survey studies [6, 40, 58, 82, 93], we present an overview of NIOA and its characteristics. Image clustering is considered an optimization issue by NIOA, and it is solved repeatedly by minimising or exploiting one or more objective functions. The general procedure of NIOA is represented as Algorithm 10.
The NIOA stopping, or terminating condition/criteria, is a parameter that specifies when an algorithm should be stopped [33, 93, 120]. For various NIOAs, determining the stopping condition is critical. A number of iterations (NIs), also known as generation, and a number of function evaluations (FEs) are two of the most prevalent types of terminating criteria. According to current research, FEs are preferred over NIs by today’s researchers. The number of iterations is always included when a constructed terminating condition is applied. Other terminating criteria include, exceeded threshold value, optimal solution reached, and the objective function is equal to zero, etc.
In literature some NIOA based crisp and fuzzy clustering have been developed which are mentioned as follows. A modified cuckoo search (CS) [34] based crisp clustering model had been developed which gave superior outcomes compared to traditional CS, PSO, Bat algorithm (BA), Firefly algorithm (FA), and other existing modified CS based clustering models. Traditional CS [31] also showed its efficient performance in the breast histology image clustering domain by outperforming classical K-means. A modified Flower Pollination Algorithm (FPA) [35] based crisp clustering model was also proposed in the pathology image segmentation domain. The suggested modified FPA outperformed various well-known NIOAs such as BA, FA, PSO, and others, according to experimental results. Stochastic Fractal Search (SFS) based crisp clustering technique had been utilized for the accurate segmentation of white blood cells (WBC) from the blood pathology images of leukaemia patients [36]. Numerical results showed that the SFS gave nonpareil outcomes to other tested NIOA and some state-of-the-art image clustering approaches. Dash et al. [29] instigated seeker optimization (SO), artificial bee colony (ABC), ant colony optimization (ACO), and particle swarm optimization (PSO) based on crisp and fuzzy clustering techniques for the optimal lesion segmentation and established satisfactory outcomes. In [76, 78], researchers also announced the NIOA with K-means to implement proper clustering-based image segmentation practices.
NIOA are also employed in fuzzy clustering domain for image segmentation. For example, Tongbram et al. [136] developed whale optimizer based fuzzy clustering model which had the better efficiency over conventional FCM clustering for MRI image segmentation. Vishnoi et al. [141] developed a nuclei segmentation model for breast histology by employing roulette wheel selection whale optimization (RSWOA) based fuzzy clustering. The proposed RSWOA outperformed differential evolution (DE), grey wolf optimizer (GWO), bat algorithm (BA), and grasshopper optimizer (GO) in this fuzzy image clustering domain. Narmatha et al. [107] developed a brain-storm optimization algorithm based FCM for the precise tumor region segmentation from MRI images. The proposed technique gave competitive results compare to PSO, glow swarm optimization (GSO), and whale swarm optimization (WSO) based clustering models. Tiwari and Jain [135] proposed an exponential grasshopper optimization algorithm-based fuzzy clustering model for histopathological image segmentation. The simulation results demonstrated the efficiency of the proposed one over other tested classical clustering techniques such as K-means and fuzzy c-means. Fred et al. [47] proposed a crow search algorithm (CSA) based FCM for the accurate segmentation of abdomen CT images. The proposed clustering model provided better segmented outcomes compared to Artificial Bee Colony (ABC), Firefly, Cuckoo, Cuckoo Search (CS), and Simulated Annealing (SA) based FCM models. A comparative study among NIOA based KM and NIOA based FCM had been done for skin lesion segmentation by Dash et al. [57]. Experimental results showed that Seeker optimization (SO) algorithm with FCM gave superior results to SO with KM to segment the lesion optimally and SO also provided competitive results compare to ACO, ABC, and PSO. For noisy image segmentation, Rapaka et al. [119] developed a morphological reconstruction fuzzy c-means clustering (MRFCM) based on an improved differential search algorithm for iris image segmentation where morphological reconstruction incorporates the power of noise immunity. The proposed technique outperformed improved particle swarm optimization based morphological reconstruct fuzzy c-means (IPSO-MRFCM), PSO-FCM in terms segmentation accuracy. Depending on the cooperation principle, Abdellahoum et al. [2] developed a Cooperative System using Fuzzy C-Means (CSFCM) methodology by using Biogeography Based Optimization (BBO), Genetic Algorithm (GA), and Firefly Algorithm (FA). Experimental results showed that the CSFCM provided good segmentation result by outperforming other tested algorithms such as PSO and ABC.
There are other ways where NIOA and KM or FCM can be hybridized to develop efficient clustering strategies. The effectiveness and superiority of one NIOA over another is problem dependent, according to the NFL theorem. However, it is also true that the algorithm design perspective has a significant impact on the effectiveness of any algorithm, and NIOA are no exception. As a result, it is clear that some NIOA are flawed in terms of basic abilities such as individual mixing and diversity, a best balance between exploration and exploitation search abilities, and so on. The trade-off between exploration/global search and exploitation/local search affects every NIOA technique. Randomization in the NIOA essentially aids them in performing a global search. As a result, because KM or FCM are very efficient local optimizers and greedy by nature, they can be included into NIOA as a local search component during clustering [40, 113]. As a result, a well-balanced exploration and exploitation is provided by randomization-based global search and traditional clustering algorithm-based local search for the NIOA.
As an instance, Hrosik et al. [66] created an enhanced image clustering strategy based on the Firefly algorithm, where the k-means clustering algorithm improves the Firefly algorithm’s results. Experiments indicated that the suggested strategy outperformed traditional KM and NIOA-based clustering algorithms.
Again, the combination of NIOA and KM/FCM can be performed in other way. The global optimal solution discovered by NIOA can be used to start the KM/ FCM algorithm. This concept can help to solve the problem of local trapping caused by random centre initialization. As an instance Li et al. [88] proposed a PSO-based K-Means algorithm for image segmentation, which used the global optimal solution to initialize the K-Means algorithm. Experimental results shows that the approach overcomes the issue of easy falling into local optimum and produced excellent outcomes.
Incorporating the output solutions of the K-means algorithm as input to NIOA is another technique to improve the performance of the algorithm. For example, Nanda et al. [106] improved the efficiency of Galactic Swarm Optimization (GSO) by using the K-means algorithm’s resultant solutions as input of GSO. The K-Means method is run for a set number of iterations in this hybrid approach, and the outcome is used as the GSO’s population’s starting solution. All the above discussed techniques are pixel-based clustering. Some histogram-based clustering techniques are developed in [15, 27, 39, 85, 120, 132] and they gave satisfactory results within less time.
5 Experimental results
The experimental study has been carried out over 100 Rosette plant images and 100 Oral histopathology images using MatlabR2018b and Windows-10 OS, ×64-based PC, Intel core i5 CPU with 8 GB RAM. The following two dataset is used in this study.
-
1)
The Rosette plant images are collected from Computer Vision Problems in Plant Phenotyping (CVPPP) benchmark datasets [99] and the web-link is: https://download.fz-juelich.de/ibg-2/Plant_Phenotyping_Datasets.zip. A1, A2 and A3 datasets are include in benchmark dataset. A1 contains RGB images of wild-type Arabidopsis plants. A2 contains RGB images of four distinct Arabidopsis mutants with varying leaf forms and sizes. A3 dataset consists of tobacco plant images. The genotype, background appearance, and composition of these datasets vary due to the different experimental setups used. The images in each dataset have various dimensions 500 × 530 pixels in A1, 530 × 565 in A2 and 2448 × 2048 pixels in A3.
-
2)
The Oral histopathology images are collected from [117] and the weblink is: https://data.mendeley.com/datasets/ftmp4cvtmb/1. It is the most widely used dataset for oral histopathology images. The dataset contains total 1224 images. The images are split into two groups, each with a different resolution. The first collection includes 89 histological images of normal oral epithelium and 439 images of Oral Squamous Cell Carcinoma (OSCC) at a magnification of 100x. The second collection includes 201 images of normal oral cavity epithelium and 495 histopathological images of OSCC at 400x magnification. The images were taken with a Leica ICC50 HD microscope from H&E-stained tissue slides from 230 patients. The slides were collected, prepared, and catalogued by medical experts.
In the field of agriculture plant growth is the major component that is why plant image analysis play an important role. It allows for the frequent and precise recording of morphological plant features. The growth of the plant analysed by the deeply rely on the leaves and their segmented image. The Rosette plant images are collected from [99], which contain the original images as well as the corresponding ground truth images.
Oral cancer is now a days a common cancer in the world. As per Oral clinicians, it is established that the Oral submucous fibrosis (OSF) initially originates and propagates in the epithelial layer. So, more accurate segmentation of this layer is extremely for clinician to make a diagnostic decision. That is why Oral histopathology plays a very important role to diagnosing the oral cancer. The Oral histopathology images collected from [117]. It consists of total 528 images; out of which of 89 are histopathological images with the normal epithelium of the oral cavity and 439 images are in Oral Squamous Cell Carcinoma (OSCC) category in 100x magnification.
For pre-processing task, the Simple Linear Iterative Clustering (SLIC) is employed to generate superpixels. The four most often used clustering algorithms, namely K-Means (KM), Fuzzy C-Means (FCM), Genetic Algorithm (GA) and Particle Swarm Optimization (PSO) has been used for leaf segmentation of Rosette plant images and epithelium layer of Oral histopathology images. A comparative study is done among the above clustering techniques and their performance both with and without superpixel pre-processing. The following are the parameters for the corresponding clustering techniques:
K is the desired number of roughly equal-sized superpixels, takes as input for SLIC algorithm. The approximate size of each super-pixel in an image of N pixels is N/K pixels. Every grid interval would have a superpixel centre for nearly equal-sized superpixels \( S=\sqrt{N/K} \). Number of superpixel used to segment the input image (K) = 2000 is set, which is optimally set from the experience. The user determines the number of cluster prototypes, which is set at 3 for all clustering techniques. The KM algorithm is stopped, if the change in centroid values is smaller than η . For FCM, the fuzzification parameter is set to 2, and the procedure is stopped if the largest difference between two successive partition matrices U is lesser than the minimal error threshold η. Mathematically, if [Max{Ut − Ut + 1}] < η then stop, where minimal error threshold η = 10−5.The probabilities of crossover and mutation, the population size, and the number of generations are all factors that are included in Genetic Algorithm. For PSO, the acceleration factors c1 and c2 control the impact of the best local and global solutions on the existing solution. In the experiment both c1 and c2 are set to 2. The experiment was conducted with a population size (n) = 50. The number of function evaluations NFE = 500 × d is taken into account for each execution of the tested objective function, as the optimization process’s stop condition, where d is the number of clusters. In an optimization problem formulation, this refers to the d-dimensional search space.
5.1 Performance evaluation parameters
To ensure a method’s validity, it must be evaluated in terms of its performance. The performance of the utilized clustering techniques has been evaluated by calculating four ground truth-based performance evaluation parameters which are summarized in Table 4. Here, TP - true positive, FP - false positive, TN - true negative, FN - false negative.
In digital imaging processing, the image performance evaluation parameters play a crucial role. Therefore, it’s vital to look at the accuracy, precision and other parameters of the clusters created by the methodologies used. It is also important to examine whether the concerned clustering techniques give the higher similarities between the data objects of same cluster and give lower similarities between the data object which are not in the same cluster. Quality evaluation parameters are basically two types, one is Ground Truth (GT) based quality parameters and another one is Referenced or Original image-based quality parameters.
-
1)
Ground Truth (GT) based quality parameters: Ground truth-based image segmentation quality parameters namely Segmentation Accuracy (SA) [55], Precision Index (PI) [55], Specitivity Index (SI) [55], Recall Index (RI) [55], Dice Index (DI) [55], Jaccard Index (JI) [55], Matthews Correlation Coefficient (MCC) [55], Comparison Score (CS) [101], Boundary Displacement Error (BDE) [101], Probability Rand Index (PRI) [101], Variation of Information (VoI) [101], and Global Consistency Error (GCE) [101] are the techniques for represent the quality of the segmented image. For labelling information of the pixel, these parameters are use.
-
2)
Referenced or Original image-based quality parameters: Referenced or Original image-based quality parameters namely Feature Similarity Index (FSIM) [28, 101], Root Mean Squared Error (RMSE) [101], Peak Signal to Noise Ratio (PSNR) [28, 101], Structural Similarity Index (SSIM) [101], Normalized Cross-Correlation (NCC) [101], Average Difference (AD), Maximum Difference (MD) [101], Normalized Absolute Error (NAE) [101]. SSIM and FSIM are two of these measures that consider perceived image quality and quantitatively analyse the segmentation results against the consequence of human observers. Instead of using pixel labelling information, these parameters employ pixel information.
5.2 Results and discussion over rosette plant images
KM, FCM, GA and PSO, the four well known clustering algorithm have been utilized to segment the leaves of the rosette plant images. For superpixel creation, SLIC method is used. Firstly, the images are segmented using four simple clustering technique and that has been compared with the superpixel based image clustering technique. The performance between simple clustering and clustering with superpixel pre-processing is discuss in this section. The above clustering method also compared with SFFCM algorithm. Fig. 6 represents the original color rosette plant image and their segmented leaf part by the four utilized algorithms with and without superpixel pre-processing. Figure 7 demonstrates the ground truth images of the leaf segmentation provided by the experts and the binary segmented leaf part provided by the employed clustering algorithms. The visual analysis of the both Figs. 6 and 7 clearly show that the SFFCM provides the best leaf-based segmentation results. Not only visual analysis, the segmentation efficacy of the clustering algorithms has been analysed by computing four well-known segmentation quality parameters namely Accuracy, MCC, Dice and Jaccard. The average values of the segmentation quality parameters over 100 images are given in Table 5. The best numerical values of the Table 5 are given in bold. Most of the values of the quality parameters clearly reveal that SFFCM provides superior outcomes over other clustering algorithms. The graphical representation of the average quality parameters (recorded in Table 5) is also showed in Fig. 8. The average execution times of the four clustering algorithms over 100 images are also presented in Table 5.
It is clearly noticed that use of superpixel as a pre-processing task decrease overall computational time. As an example, for plant images the computational time of PSO algorithm is 14.30 sec but use of superpixel decrease the PSO computational which is 13.73 sec.
5.3 Result and discussion over Oral histopathology images
The four well known clustering algorithm, namely KM, FCM, GA and PSO have been utilized to segment the epithelium layer of Oral histopathology images. For superpixel creation, SLIC method is used. Firstly, the images are segmented using four simple clustering technique and that has been compared with the superpixel based image clustering technique. The performance between simple clustering and clustering with superpixel pre-processing is discuss in this section. The above clustering method also compared with SFFCM algorithm. Figure 9 represents the original color Oral histopathology images and their segmented epithelium layer part by the four utilized algorithms with and without superpixel pre-processing. Figure 10 demonstrates the ground truth images of the epithelium layer segmentation provided by the experts and the binary segmented epithelium layer part provided by the employed clustering algorithms. The visual analysis of the both Figs. 9 and 10 clearly show that the SFFCM provides the best epithelium layer segmentation results. Not only visual analysis, the segmentation efficacy of the clustering algorithms has been analysed by computing four well-known segmentation quality parameters namely Accuracy, MCC, Dice and Jaccard. The average values of the segmentation quality parameters over 100 images are given in Table 6. The best numerical values of the Table 6 are given in bold. Most of the values of the quality parameters clearly reveal that SFFCM provides superior outcomes over other clustering algorithms. The graphical representation of the average quality parameters (recorded in Table 6) is also showed in Fig. 11. The average execution times of the four clustering algorithms over 100 images are also presented in Table 6.
For Oral histopathology images the execution time of PSO and SLIC-PSO is 14.75 sec and 14.44 sec respectively. It is clearly noticed that use of SLIC pre-processing technique reduce the overall computation time. As the number of superpixel used to segment the input image (K) value depends on user the time complexity for superpixel generation may vary.
6 Conclusion and future directions
Image segmentation is always a demanding task but producing segmented image with an ideal time is very challenging. So, minimizing pixel quantity can decrease the computation time. Another significant advantage of superpixel based image segmentation is the noise immunity capability. In this situation, superpixels are become increasingly popular in many computers vision application. Therefore, this study provides an in-depth review on the utilization of the superpixel images for accurate image segmentation while employed with clustering techniques. The literature survey of the past five years has been presented in Table 1. Then the brief discussion on the recent superpixel generation techniques has also been presented in the paper. However, it is found that selecting the right superpixel algorithm and its parameters for the particular application are crucial. But, out of many superpixel algorithms, SLIC is the mostly used superpixel generation algorithm in the published research works. Clustering is a simple but effective image segmentation procedure. Hence, this study concentrates on the measuring their efficacy while employed over superpixel images. A depth presentation and discussion on the well-known clustering techniques and their pros-cons have been performed here. The discussion demonstrates that the ideal dataset for hierarchical clustering is arbitrary shape and attribute of arbitrary type. Hierarchical clustering algorithms create a hierarchy of data pieces, they are suitable for both convex and arbitrary data. Partitional clustering technique is mostly used. Because the image is separated on the basis of different features, the image segmentation approach can be employed according to the application or usage. According to some pre-set objective functions, partitional clustering algorithms produce disjoint clusters. Because of their excellent computing efficiency and low time complexity, partitional clustering algorithms are recommended. Partitional clustering methods are chosen over other clustering approaches for large datasets. Despite the fact that partitional clustering is computationally efficient, the number of clusters must be defined advance. Furthermore, partitional clustering algorithms are sensitive to outliers and can trap local optima in some cases. However, choosing the right clustering algorithm basically depends on certain parameters such as type of data, scalability, sensitivity towards noise and number of clusters.
At last, this study performs a comparative study among the four widely used clustering techniques namely K-Means (KM), Fuzzy C-Means (FCM), Genetic Algorithm (GA) and Particle Swarm Optimization (PSO) are used for segmenting leaves of Rosette plant images and Epithelial layer of Oral histological images. For superpixel generation, one of the most prominent methods, Simple Linear Iterative Clustering (SLIC), is used. These techniques are also compared to SFFCM algorithm which used MMGR-WT for producing superpixel image. In terms of visual and numerical analysis of the used segmentation quality parameters, experimental results show that SFFCM gives the best results for both Rosette plant images and Oral histopathology images. For Rosette plant images, segmentation accuracy of the SFFCM is 96.64%. Whereas, KM, SLIC-KM, FCM, SLIC-FCM, SLIC-GA, PSO, SLIC-PSO associate with 96.11%, 95.99%, 96.59%, 96.42%, 96.21%, 96.61%, and 96.59% segmentation accuracy respectively. For Oral images, segmentation accuracy of the SFFCM is 98.68%. Whereas, KM, SLIC-KM, FCM, SLIC-FCM, SLIC-GA, PSO, SLIC-PSO associate with 94.70%, 96.72%, 94.61%, 95.99%, 94.65%, 94.63% and 96.82% segmentation accuracy respectively. Based on the mentioned findings, we infer that superpixel image-based clustering algorithms perform better in terms of segmentation accuracy and other quality parameters than typical clustering-based algorithms. Superpixel techniques add a compact restriction to the objectness function, resulting more compact, coherent, and regular superpixels. The trade-off between speed and accuracy in superpixel image-based segmentation is worthwhile to examine. The time it takes for certain algorithms to run is mostly determined by the number of iterations they go through during optimization.
It is our assumption that this study will be valuable to researchers working on superpixel segmentation, clustering with superpixel images and related topics and also their application. Several future directions can be found depending on this analytical review paper and are listed as follows.
The main future work which can be done based on only superpixel generation techniques is the utilization of proper superpixel generation technique for different kinds of images which is fully automatic i.e., there is no need of parameter tuning including the selection of number of superpixels. In addition to the above future work, incorporation of Deep learning in superpixel based image segmentation can be a bright future direction.
For clustering techniques like KM and FCM, the four major future directions are: (i) Selection of proper cluster number in prior to clustering automatically. (ii) Enhancement of their clustering efficacy for the noisy images especially without knowing the noise-type. (iii) Development of proper centres initialization techniques so that they do not converge prematurely. On the other hand, the local trapping problem of KM or FCM can be overcome by some future strategies so that they can able to find the actual or nearly global optima. (iv) The computational time of the KM and FCM for image segmentation significantly depends on the image size i.e., number of pixels. Therefore, finding image size independent fast, accurate, and noise-robust clustering techniques will be an emerging future direction.
Data availability
My manuscript has no associated data.
References
Abd Elaziz M, Abo Zaid EO, Al-qaness MA, Ibrahim RA (2021) Automatic Superpixel-based clustering for color image segmentation using q-generalized Pareto distribution under linear normalization and hunger games search. Mathematics 9(19):2383. https://doi.org/10.3390/math9192383
Abdellahoum H, Mokhtari N, Brahimi A, Boukra A (2021) CSFCM: an improved fuzzy C-means image segmentation algorithm using a cooperative approach. Expert Syst Appl 166:114063. https://doi.org/10.1016/j.eswa.2020.114063
Achanta R, Susstrunk S (2017) Superpixels and polygons using simple non-iterative clustering. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4651–4660. https://doi.org/10.1109/CVPR.2017.520
Achanta R, Shaji A, Smith K, Lucchi A, Fua P, Süsstrunk S (2010) Slic superpixels (No. REP_WORK)
Achanta R, Shaji A, Smith K, Lucchi A, Fua P, Süsstrunk S (2012) SLIC superpixels compared to state-of-the-art superpixel methods. IEEE Trans Pattern Anal Mach Intell 34(11):2274–2282. https://doi.org/10.1109/tpami.2012.120
Ahmed MN, Yamany SM, Mohamed N, Farag AA, Moriarty T (2002) A modified fuzzy c-means algorithm for bias field estimation and segmentation of MRI data. IEEE Trans Med Imaging 21(3):193–199. https://doi.org/10.1109/42.996338
Albayrak A, Bilgin G (2019) Automatic cell segmentation in histopathological images via two-staged superpixel-based algorithms. Med Biol Eng Comput 57(3):653–665. https://doi.org/10.1007/s11517-018-1906-0
Anter AM, Hassenian AE (2019) CT liver tumor segmentation hybrid approach using neutrosophic sets, fast fuzzy c-means and adaptive watershed algorithm. Artificial Intell Med 97:105–117. https://doi.org/10.1016/j.artmed.2018.11.007
Arbelaez P, Maire M, Fowlkes C, Malik J (2010) Contour detection and hierarchical image segmentation. IEEE Trans Pattern Anal Mach Intell 33(5):898–916. https://doi.org/10.1109/tpami.2010.161
Armato SG III, McLennan G, McNitt-Gray MF, Meyer CR, Yankelevitz D, Aberle DR, Clarke LP (2004) Lung image database consortium: developing a resource for the medical imaging research community. Radiology 232(3):739–748. https://doi.org/10.1148/radiol.2323032035
Benesova W, Kottman M (2014) Fast superpixel segmentation using morphological processing. In: In Conference on Machine Vision and Machine Learning, pp 67–61
Bezdek JC, Ehrlich R, Full W (1984) FCM: the fuzzy c-means clustering algorithm. Comput Geosci 10(2–3):191–203. https://doi.org/10.1016/0098-3004(84)90020-7
Bouguettaya A, Yu Q, Liu X, Zhou X, Song A (2015) Efficient agglomerative hierarchical clustering. Expert Syst Appl 42(5):2785–2797. https://doi.org/10.1016/j.eswa.2014.09.054
Buyssens P, Gardin I, Ruan S (2014) Eikonal based region growing for superpixels generation: application to semi-supervised real time organ segmentation in CT images. Irbm 35(1):20–26. https://doi.org/10.1016/j.irbm.2013.12.007
Cai W, Chen S, Zhang D (2007) Fast and robust fuzzy c-means clustering algorithms incorporating local information for image segmentation. Pattern Recogn 40(3):825–838. https://doi.org/10.1016/j.patcog.2006.07.011
Celebi ME, Wen Q, Hwang S (2015) An effective real-time color quantization method based on divisive hierarchical clustering. J Real-Time Image Proc 10(2):329–344. https://doi.org/10.1007/s11554-012-0291-4
Chakraborty S, Mali K (2021) SuFMoFPA: a superpixel and meta-heuristic based fuzzy image segmentation approach to explicate COVID-19 radiological images. Expert Syst Appl 167:114142. https://doi.org/10.1016/j.eswa.2020.114142
Chavent M, Lechevallier Y, Briant O (2007) DIVCLUS-T: a monothetic divisive hierarchical clustering method. Comput Stat Data Anal 52(2):687–701. https://doi.org/10.1016/j.csda.2007.03.013
Chen S, Zhang D (2004) Robust image segmentation using FCM with spatial constraints based on new kernel-induced distance measure. IEEE Trans Syst Man Cybern B Cybern 34(4):1907–1916. https://doi.org/10.1109/tsmcb.2004.831165
Chen J, Li Z, Huang B (2017) Linear spectral clustering superpixel. IEEE Trans Image Process 26(7):3317–3330. https://doi.org/10.1109/tip.2017.2651389
Cheng MM, Mitra NJ, Huang X, Torr PH, Hu SM (2014) Global contrast based salient region detection. IEEE Trans Pattern Anal Mach Intell 37(3):569–582. https://doi.org/10.1109/tpami.2014.2345401
Chouhan SS, Singh UP, Jain S (2020) Applications of computer vision in plant pathology: a survey. Arch Comput Methods Eng 27(2):611–632. https://doi.org/10.1007/S11831-019-09324-0
Comaniciu D, Meer P (2002) Mean shift: a robust approach toward feature space analysis. IEEE Trans Pattern Anal Mach Intell 24(5):603–619. https://doi.org/10.1109/34.1000236
Cong L, Ding S, Wang L, Zhang A, Jia W (2018) Image segmentation algorithm based on superpixel clustering. IET Image Process 12(11):2030–2035. https://doi.org/10.1049/iet-ipr.2018.5439
Conrad C, Mertz M, Mester R (2013) Contour-relaxed superpixels. In: International workshop on energy minimization methods in computer vision and pattern recognition. Springer, Berlin, Heidelberg, pp 280–293. https://doi.org/10.1007/978-3-642-40395-8_21
Das S, Konar A, Chakraborty UK (2006) Automatic fuzzy segmentation of images with differential evolution. IEEE Congress on Evolutionary Computation 2006:2026–2033. https://doi.org/10.1109/CEC.2006.1688556
Das A, Dhal KG, Ray S, Gálvez J (2021) Histogram based fast and robust image clustering using stochastic fractal search and morphological reconstruction. Neural Comput & Applic 34:4531–4554. https://doi.org/10.1007/s00521-021-06610-6
Das A, Namtirtha A, Dutta A (2022) Fuzzy clustering of acute lymphoblastic leukemia images assisted by eagle strategy and morphological reconstruction. Knowl-Based Syst 239:108008. https://doi.org/10.1016/j.knosys.2021.108008
Dash M, Londhe ND, Ghosh S, Shrivastava VK, Sonawane RS (2020) Swarm intelligence based clustering technique for automated lesion detection and diagnosis of psoriasis. Comput Biol Chem 86:107247. https://doi.org/10.1016/j.compbiolchem.2020.107247
Dhal KG, Fister I Jr, Das A, Ray S, Das S (2018) Breast histopathology image clustering using cuckoo search algorithm. In: Proceedings of the 5th student computer science research conference, pp 47–12
Dhal KG, Fister I Jr, Das A, Ray S, Das S (2018) Breast histopathology image clustering using cuckoo search algorithm. In: 5th Student Computer Science Research Conference, vol 2018. University of Maribor, Slovenia, pp 47–54
Dhal KG, Ray S, Das A, Das S (2019) A survey on nature-inspired optimization algorithms and their application in image enhancement domain. Arch Comput Methods Eng 26(5):1607–1638. https://doi.org/10.1007/S11831-018-9289-9
Dhal KG, Ray S, Das A, Gálvez J, Das S (2019) Fuzzy multi-level color satellite image segmentation using nature-inspired optimizers: a comparative study. J Indian Soc Remote Sens 47(8):1391–1415. https://doi.org/10.1007/s12524-019-01005-6
Dhal KG, Das A, Ray S, Das S (2019) A clustering based classification approach based on modified cuckoo search algorithm. Pattern Recognit Image Anal 29(3):344–359. https://doi.org/10.1134/S1054661819030052
Dhal KG, Gálvez J, Das S (2019) Toward the modification of flower pollination algorithm in clustering-based image segmentation. Neural Comput & Applic:1–19. https://doi.org/10.1007/s00521-019-04585-z
Dhal KG, Gálvez J, Ray S, Das A, Das S (2020) Acute lymphoblastic leukemia image segmentation driven by stochastic fractal search. Multimed Tools Appl 79:12227–12255. https://doi.org/10.1007/s11042-019-08417-z
Dhal KG, Das A, Gálvez J, Ray S, Das S (2020) An overview on nature-inspired optimization algorithms and their possible application in image processing domain. Pattern Recognit Image Anal 30(4):614–631. https://doi.org/10.4018/IJAMC.292516
Dhal KG, Das A, Ray S, Gálvez J, Das S (2020) Nature-inspired optimization algorithms and their application in multi-thresholding image segmentation. Arch Comput Methods Eng 27(3):855–888. https://doi.org/10.1007/S11831-019-09334-Y
Dhal KG, Das A, Ray S, Gálvez J (2021) Randomly attracted rough firefly algorithm for histogram based fuzzy image clustering. Knowl-Based Syst 216:106814. https://doi.org/10.1016/j.knosys.2021.106814
Dhal KG, Das A, Ray S, Sarkar K, Gálvez J (2021) An analytical review on rough set based image clustering. Arch Comput Methods Eng:1–30. https://doi.org/10.1007/s11831-021-09629-z
Dhal KG, Das A, Ray S, Gálvez J, Das S (2021) Histogram equalization variants as optimization problems: a review. Arch Comput Methods Eng 28(3):1471–1496. https://doi.org/10.1007/s11831-020-09425-1
Dhillon IS, Mallela S, Kumar R (2003) A divisive information theoretic feature clustering algorithm for text classification. J Mach Learn Res 3:1265–1287
Drucker F, MacCormick J (2009, December) Fast superpixels for video analysis. In: 2009 Workshop on Motion and Video Computing (WMVC). IEEE. pp. 1–8
Eberhart R, Kennedy J (1995) A new optimizer using particle swarm theory. In: Micro Machine and Human Science, 1995. MHS'95. Proceedings of the Sixth International Symposium on IEEE, pp 39–43. https://doi.org/10.1109/MHS.1995.494215
Elkhateeb E, Soliman H, Atwan A, Elmogy M, Kwak KS, Mekky N (2021) A novel coarse-to-Fine Sea-land segmentation technique based on Superpixel fuzzy C-means clustering and modified Chan-Vese model. IEEE Access 9:53902–53919. https://doi.org/10.1109/ACCESS.2021.3065246
Felzenszwalb PF, Huttenlocher DP (2004) Efficient graph-based image segmentation. Int J Comput Vis 59(2):167–181. https://doi.org/10.1023/B%3AVISI.0000022288.19776.77
Fred AL, Kumar SN, Padmanaban P, Gulyas B, Kumar HA (2020) Fuzzy-crow search optimization for medical image segmentation. In: Applications of hybrid metaheuristic algorithms for image processing. Springer, Cham, pp 413–439. https://doi.org/10.1007/978-3-030-40977-7_18
Fu H, Cao X, Tang D, Han Y, Xu D (2014) Regularity preserved superpixels and supervoxels. IEEE Trans Multimedia 16(4):1165–1175. https://doi.org/10.1109/TMM.2014.2305571
Fu Z, Sun Y, Fan L, Han Y (2018) Multiscale and multifeatured segmentation of high-spatial resolution remote sensing images using superpixels with mutual optimal strategy. Remote Sens 10(8):1289. https://doi.org/10.3390/rs10081289
Fumero F, Alayón S, Sanchez JL, Sigut J, Gonzalez-Hernandez M (2011) RIM-ONE: An open retinal image database for optic nerve evaluation. In: 2011 24th international symposium on computerbased medical systems (CBMS), pp 1–6. IEEE. https://doi.org/10.1109/CBMS.2011.5999143
Gao Y, Lin J, Xie J, Ning Z (2020) A real-time defect detection method for digital signal processing of industrial inspection applications. IEEE Trans Indust Inform 17(5):3450–3459. https://doi.org/10.1109/TII.2020.3013277
George Y, Aldeen M, Garnavi R (2016) Pixel-based skin segmentation in psoriasis images. In: 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). IEEE, pp 1352–1356. https://doi.org/10.1109/embc.2016.7590958
George YM, Aldeen M, Garnavi R (2017) Automatic psoriasis lesion segmentation in two-dimensional skin images using multiscale superpixel clustering. J Med Imaging 4(4):044004. https://doi.org/10.1117/1.JMI.4.4.044004
Ghaffari R, Golpardaz M, Helfroush MS, Danyali H (2020) A fast, weighted CRF algorithm based on a two-step superpixel generation for SAR image segmentation. Int J Remote Sens 41(9):3535–3557. https://doi.org/10.1080/01431161.2019.1706202
Ghosal D, Das A, Dhal KG (2020) A comparative study among clustering techniques for leaf segmentation in rosette plants. Pattern Recognit Image Anal 31(4). https://doi.org/10.1134/S1054661821040118
Giraud R, Berthoumieu Y (2019) Texture superpixel clustering from patch-based nearest neighbor matching. In: 2019 27th European Signal Processing Conference (EUSIPCO). IEEE, pp 1–5. https://doi.org/10.23919/EUSIPCO.2019.8902729
Goldberg DE, Holland JH (1988) Genetic algorithms and machine learning. Mach Learn 3(2):95–99. https://doi.org/10.1023/A%3A1022602019183
Gong M, Zhou Z, Ma J (2011) Change detection in synthetic aperture radar images based on image fusion and fuzzy clustering. IEEE Trans Image Process 21(4):2141–2151. https://doi.org/10.1109/tip.2011.2170702
Gong M, Liang Y, Shi J, Ma W, Ma J (2012) Fuzzy c-means clustering with local information and kernel metric for image segmentation. IEEE Trans Image Process 22(2):573–584. https://doi.org/10.1109/tip.2012.2219547
Goyal P, Kumari S, Sharma S, Kumar D, Kishore V, Balasubramaniam S, Goyal N (2016) A fast, scalable SLINK algorithm for commodity cluster computing exploiting spatial locality. In: 2016 IEEE 18th International Conference on High Performance Computing and Communications; IEEE 14th International Conference on Smart City; IEEE 2nd International Conference on Data Science and Systems (HPCC/SmartCity/DSS). IEEE, pp 268–275. https://doi.org/10.1109/HPCC-SmartCity-DSS.2016.0047
Gronau I, Moran S (2007) Optimal implementations of UPGMA and other common clustering algorithms. Inf Process Lett 104(6):205–210. https://doi.org/10.1016/j.ipl.2007.07.002
Guénoche A, Hansen P, Jaumard B (1991) Efficient algorithms for divisive hierarchical clustering with the diameter criterion. J Classif 8(1):5–30. https://doi.org/10.1007/BF02616245
Guha S, Rastogi R, Shim K (1998) CURE: an efficient clustering algorithm for large databases. ACM SIGMOD Rec 27(2):73–84. https://doi.org/10.1145/276304.276312
Ha NT, Manley-Harris M, Pham TD, Hawes I (2021) The use of radar and optical satellite imagery combined with advanced machine learning and metaheuristic optimization techniques to detect and quantify above ground biomass of intertidal seagrass in a New Zealand estuary. Int J Remote Sens 42(12):4712–4738. https://doi.org/10.1080/01431161.2021.1899335
Hamamci A, Unal G (2012) Multimodal brain tumor segmentation using the tumor-cut method on the BraTS dataset. Proc MICCAI-BRATS:19–23
Hrosik RC, Tuba E, Dolicanin E, Jovanovic R, Tuba M (2019) Brain image segmentation based on firefly algorithm combined with k-means clustering. Stud Inform. Control 28:167–176. https://doi.org/10.24846/V28I2Y201905
Humayun A, Li F, Rehg JM (2015) The middle child problem: Revisiting parametric min-cut and seeds for object proposals. In: Proceedings of the IEEE International Conference on Computer Vision, pp 1600–1608
Ibrahim A, El-kenawy ESM (2020) Image segmentation methods based on superpixel techniques: a survey. J Comput Sci Inf Syst 15(3)
Ilesanmi AE, Idowu OP, Makhanov SS (2020) Multiscale superpixel method for segmentation of breast ultrasound. Comput Biol Med 125:103879. https://doi.org/10.1016/j.compbiomed.2020.103879
Irshad H, Montaser-Kouhsari L, Waltz G, Bucur O, Nowak JA, Dong F, Beck AH (2014) Crowdsourcing image annotation for nucleus detection and segmentation in computational pathology: evaluating experts, automated methods, and the crowd. In: Pacific symposium on biocomputing Co-chairs, pp 294–305. https://doi.org/10.1142/9789814644730_0029
Ishizaka A, Lokman B, Tasiou M (2021) A stochastic multi-criteria divisive hierarchical clustering algorithm. Omega 103:102370. https://doi.org/10.1016/j.omega.2020.102370
Jia X, Lei T, Liu P, Xue D, Meng H, Nandi AK (2020) Fast and automatic image segmentation using Superpixel-based graph clustering. IEEE Access 8:211526–211539. https://doi.org/10.1109/ACCESS.2020.3039742
Kauppi T, Kalesnykiene V, Kamarainen JK, Lensu L, Sorri I, Raninen A, Pietilä J (2007) The diaretdb1 diabetic retinopathy database and evaluation protocol. BMVC 1(1):10. https://doi.org/10.5244/C.21.15
Karaboga D, Basturk B (2007) A powerful and efficient algorithm for numerical function optimization: artificial bee colony (ABC) algorithm. J Glob Optim 39(3):459–471. https://doi.org/10.1007/S10898-007-9149-X
Karypis G, Han E-H, Kumar V (1999) Chameleon: hierarchical clustering using dynamic modeling. Computer 32:68–75. https://doi.org/10.1109/2.781637
Kate V, Shukla P (2020) Image segmentation of breast Cancer histopathology images using PSO-based clustering technique. In: Social networking and computational intelligence. Springer, Singapore, pp 207–216. https://doi.org/10.1007/978-981-15-2071-6_17
Khosravanian A, Rahmanimanesh M, Keshavarzi P, Mozaffari S (2021) Fast level set method for glioma brain tumor segmentation based on superpixel fuzzy clustering and lattice boltzmann method. Comput Methods Prog Biomed 198:105809. https://doi.org/10.1016/j.cmpb.2020.105809
Khrissi L, El Akkad N, Satori H, Satori K (2020) Image segmentation based on k-means and genetic algorithms. In: Embedded systems and artificial intelligence. Springer, Singapore, pp 489–497. https://doi.org/10.1007/978-981-15-0947-6_46
Kim YI, Kim WH, Kim TJ, Choi KW (1992) Histopographic characterization of chronic gastritis associated with early gastric carcinomas. Korean J Gastroenterol 24(2):216–223
Kim S, Yoo CD, Nowozin S, Kohli P (2014) Image segmentation using higher-order correlation clustering. IEEE Trans Pattern Anal Mach Intell 36(9):1761–1774. https://doi.org/10.1109/TPAMI.2014.2303095
Kim DH, Cho H, Cho HC (2019) Gastric lesion classification using deep learning based on fast and robust fuzzy C-means and simple linear iterative clustering Superpixel algorithms. J Electr Eng Technol 14(6):2549–2556. https://doi.org/10.1007/s42835-019-00259-x
Krinidis S, Chatzis V (2010) A robust fuzzy local information C-means clustering algorithm. IEEE Trans Image Process 19(5):1328–1337. https://doi.org/10.1109/tip.2010.2040763
Kumar SN, Fred AL, Varghese PS (2019) Suspicious lesion segmentation on brain, mammograms and breast MR images using new optimized spatial feature based super-pixel fuzzy c-means clustering. J Digit Imaging 32(2):322–335. https://doi.org/10.1007/s10278-018-0149-9
Lei T, Jia X, Zhang Y, Liu S, Meng H, Nandi AK (2018) Superpixel-based fast fuzzy C-means clustering for color image segmentation. IEEE Trans Fuzzy Syst 27(9):1753–1766. https://doi.org/10.1109/TFUZZ.2018.2889018
Lei T, Jia X, Zhang Y, He L, Meng H, Nandi AK (2018) Significantly fast and robust fuzzy c-means clustering algorithm based on morphological reconstruction and membership filtering. IEEE Trans Fuzzy Syst 26(5):3027–3041. https://doi.org/10.1109/TFUZZ.2018.2796074
Levinshtein A, Stere A, Kutulakos KN, Fleet DJ, Dickinson SJ, Siddiqi K (2009) Turbopixels: fast superpixels using geometric flows. IEEE Trans Pattern Anal Mach Intell 31(12):2290–2297. https://doi.org/10.1109/tpami.2009.96
Li Z, Chen J (2015) Superpixel segmentation using linear spectral clustering. Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1356–1363. https://doi.org/10.1109/CVPR.2015.7298741
Li H, He H, Wen Y (2015) Dynamic particle swarm optimization and K-means clustering algorithm for image segmentation. Optik 126(24):4817–4822. https://doi.org/10.1016/j.ijleo.2015.09.127
Li X, Liu K, Dong Y (2017) Superpixel-based foreground extraction with fast adaptive trimaps. IEEE Trans Cybernetics 48(9):2609–2619. https://doi.org/10.1109/tcyb.2017.2747143
Li S, Li W, Qiu J (2017) A novel divisive hierarchical clustering algorithm for geospatial analysis. ISPRS Int J Geo Inf 6(1):30. https://doi.org/10.3390/ijgi6010030
Li H, Jia Y, Cong R, Wu W, Kwong S, Chen C (2020) Superpixel segmentation based on spatially constrained subspace clustering. IEEE Trans Indust Inform 17:7501–7512. https://doi.org/10.1109/TII.2020.3044068
Liu MY, Tuzel O, Ramalingam S, Chellappa R (2011) Entropy rate superpixel segmentation. In: CVPR 2011. IEEE, pp 2097–2104. https://doi.org/10.1109/CVPR.2011.5995323
Liu G, Zhao Z, Zhang Y (2015) Image fuzzy clustering based on the region-level Markov random field model. IEEE Geosci Remote Sens Lett 12(8):1770–1774. https://doi.org/10.1109/LGRS.2015.2425225
Liu Y, Wang H, Chen Y, Wu H, Wang H (2020) A passive forensic scheme for copy-move forgery based on superpixel segmentation and K-means clustering. Multimed Tools Appl 79(1):477–500. https://doi.org/10.1007/s11042-019-08044-8
Machairas E, Decencière T (2014) Walter, Waterpixels: Superpixels based on the watershed transformation. In: International Conference on Image Processing, pp 4343–4347. https://doi.org/10.1109/ICIP.2014.7025882
Maniezzo ACMDV (1992) Distributed optimization by ant colonies. In: Toward a practice of autonomous systems: proceedings of the First European Conference on Artificial Life. MIT Press, p 134
Maruthamuthu A (2020) Brain tumour segmentation from MRI using superpixels based spectral clustering. Journal of King Saud University-Computer and Information Sciences 32(10):1182–1193. https://doi.org/10.1016/j.jksuci.2018.01.009
Meyer F (2012) The watershed concept and its use in segmentation: a brief history. arXiv preprint arXiv:1202.0216. https://doi.org/10.48550/arXiv.1202.0216
Minervini M, Fischbach A, Scharr H, Tsaftaris SA (2016) Finely-grained annotated datasets for image-based plant phenotyping. Pattern Recogn Lett 81:80–89. https://doi.org/10.1016/j.patrec.2015.10.013
Mittal H, Saraswat M (2018) An image segmentation method using logarithmic kbest gravitational search algorithm based superpixel clustering. Evol Intel:1–13. https://doi.org/10.1007/s12065-018-0192-y
Mittal H, Saraswat M (2018) An optimum multi-level image thresholding segmentation using non-local means 2D histogram and exponential Kbest gravitational search algorithm. Eng Appl Artif Intell 71:226–235. https://doi.org/10.1016/j.engappai.2018.03.001
Mittal H, Saraswat M (2019) An automatic nuclei segmentation method using intelligent gravitational search algorithm based superpixel clustering. Swarm Evol Comput 45:15–32. https://doi.org/10.1016/j.swevo.2018.12.005
Mittal H, Pandey AC, Saraswat M, Kumar S, Pal R, Modwel G (2021) A comprehensive survey of image segmentation: clustering methods, performance parameters, and benchmark datasets. Multimed Tools Appl:1–26. https://doi.org/10.1007/s11042-021-10594-9
Mohamed NA, Zulkifley MA, Zaki WMDW, Hussain A (2019) An automated glaucoma screening system using cup-to-disc ratio via simple linear iterative clustering superpixel approach. Biomed Signal Process Control 53:101454. https://doi.org/10.1016/J.BSPC.2019.01.003
Murtagh F, Contreras P (2011) Methods of hierarchical clustering. arXiv preprint arXiv:1105.0121. https://doi.org/10.48550/arXiv.1105.0121
Nanda SJ, Gulati I, Chauhan R, Modi R, Dhaked U (2019) A K-means-galactic swarm optimization-based clustering algorithm with Otsu’s entropy for brain tumor detection. Appl Artif Intell 33(2):152–170. https://doi.org/10.1080/08839514.2018.1530869
Narmatha C, Eljack SM, Tuka AARM, Manimurugan S, Mustafa M (2020) A hybrid fuzzy brain-storm optimization algorithm for the classification of brain tumor MRI images. J Ambient Intell Humaniz Comput:1–9. https://doi.org/10.1007/s12652-020-02470-5
Neubert P, Protzel P (2014) Compact watershed and preemptive SLIC: on improving trade-offs of superpixel segmentation algorithms. In: International Conference on Pattern Recognition, pp 996–1001. https://doi.org/10.1109/ICPR.2014.181
Neubert P, Protzel P (2014) Compact watershed and preemptiveslic: on improving trade-offs of superpixel segmentation algorithms. In: 2014 22nd International Conference on Pattern Recognition, IEEE, pp 996–1001. https://doi.org/10.1109/ICPR.2014.181
Niharika E, Adeeba H, Krishna ASR, Yugander P (2017) K-means based noisy SAR image segmentation using median filtering and Otsu method. In: 2017 International Conference on IoT and Application (ICIOT), vol 1–4. IEEE. https://doi.org/10.1109/ICIOTA.2017.8073630
Omran M, Engelbrecht AP, Salman A (2005) Particle swarm optimization method for image clustering. Int J Pattern Recognit Artif Intell 19(03):297–321. https://doi.org/10.1142/S0218001405004083
Özdemir D, Akarun L (2002) A fuzzy algorithm for color quantization of images. Pattern Recogn 35(8):1785–1791. https://doi.org/10.1016/S0031-3203(01)00170-4
Pakhira MK (2015) A fast k-means algorithm using cluster shifting to produce compact and separate clusters. Int J Eng 28(1):35–43. https://doi.org/10.5829/idosi.ije.2015.28.01a.05
Patel S, Kadhiwala B (2018, May) Comparative Analysis of Cluster Based Superpixel Segmentation Techniques. In: 2018 2nd International Conference on Trends in Electronics and Informatics (ICOEI). IEEE, pp 1454–1459. https://doi.org/10.1109/ICOEI.2018.8553834
Potenza F, Rinaldi C, Ottaviano E, Gattulli V (2020) A robotics and computer-aided procedure for defect evaluation in bridge inspection. J Civ Struct Heal Monit 10(3):471–484. https://doi.org/10.1007/s13349-020-00395-3
Price KV (1999) An introduction to differential evolution. In: New ideas in optimization. McGraw-Hill Ltd, pp 79–108
Rahman TY, Mahanta LB, Das AK, Sarma JD (2020) Histopathological imaging database for oral cancer analysis. Data in Brief 29:105114. https://doi.org/10.1016/j.dib.2020.105114
Randen T, Husoy JH (1999) Filtering for texture classification: A comparative study. IEEE Trans Pattern Anal Mach Intell 21(4):291–310. https://doi.org/10.1109/34.761261
Rapaka S, Kumar PR, Katta M, Lakshminarayana K, Kumar NB (2021) A new segmentation method for non-ideal iris images using morphological reconstruction FCM based on improved DSA. SN Appl Sci 3(1):1–15. https://doi.org/10.1007/s42452-020-04110-1
Ray S, Das A, Dhal KG, Gálvez J, Naskar PK (2021) Cauchy with whale optimizer based eagle strategy for multi-level color hematology image segmentation. Neural Comput & Applic 33(11):5917–5949. https://doi.org/10.1007/s00521-020-05368-7
Rela M, Rao SN, Patil RR (2020) Liver tumor segmentation using superpixel based fast fuzzy C means clustering. Int J Adv Comput Sci Appl 11(11). https://doi.org/10.14569/IJACSA.2020.0111149
Ren X, Malik J (2003, October) Learning a classification model for segmentation. In: Computer Vision, IEEE International Conference on, vol 2. IEEE Computer Society, pp 10–10. https://doi.org/10.1109/ICCV.2003.1238308
Rottensteiner F, Sohn G, Jung J, Gerke M, Baillard C, Benitez S, Breitkopf U (2012) The ISPRS benchmark on urban object classification and 3D building reconstruction. ISPRS Annals of the Photogrammetry, Remote Sensing and Spatial Information Sciences I–3(1):293–298. https://doi.org/10.5194/isprsannals-I-3-293-2012
Saxena A, Prasad M, Gupta A, Bharill N, Patel OP, Tiwari A, Lin CT (2017) A review of clustering techniques and developments. Neurocomputing 267:664–681. https://doi.org/10.1016/j.neucom.2017.06.053
Sharma S, Batra N (2019) Comparative study of single linkage, complete linkage, and ward method of agglomerative clustering. In: 2019 International Conference on Machine Learning, Big Data, Cloud and Parallel Computing (COMITCon). IEEE, pp 568–573. https://doi.org/10.1109/COMITCon.2019.8862232
Shi J, Malik J (2000) Normalized cuts and image segmentation. IEEE Trans Pattern Anal Mach Intell 22(8):888–905. https://doi.org/10.1109/34.868688
Shi J, Yan Q, Xu L, Jia J (2015) Hierarchical image saliency detection on extended CSSD. IEEE Trans Pattern Anal Mach Intell 38(4):717–729. https://doi.org/10.1109/TPAMI.2015.2465960
Siyuan R, Xinying L (2020) Superpixel image segmentation based on improved K-means. J Phys Conf Ser 1533(3):032067. https://doi.org/10.1088/1742-6596/1533/3/032067
Soltani A, Battikh T, Jabri I, Lakhoua N (2018) A new expert system based on fuzzy logic and image processing algorithms for early glaucoma diagnosis. Biomed Signal Process Control 40:366–377. https://doi.org/10.1016/j.bspc.2017.10.009
Stutz D, Hermans A, Leibe B (2018) Superpixels: an evaluation of the state-of-the-art. Comput Vis Image Underst 166:1–27. https://doi.org/10.1016/j.cviu.2017.03.007
Suckling JP (1994) The mammographic image analysis society digital mammogram database. Digital Mammo:375–386 And for brain images - http://www.oasis-brains.org/
Szilagyi L, Benyo Z, Szilágyi SM, Adam HS (2003) MR brain image segmentation using an enhanced fuzzy c-means algorithm. In: Proceedings of the 25th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (IEEE Cat. No. 03CH37439), vol 1. IEEE, pp 724–726. https://doi.org/10.1109/IEMBS.2003.1279866
Tang D, Fu H, Cao X (2012) Topology preserved regular superpixel. In: 2012 IEEE International Conference on Multimedia and Expo. IEEE, pp 765–768. https://doi.org/10.1109/ICME.2012.184
Tasli HE, Cigla C, Alatan AA (2015) Convexity constrained efficient superpixel and supervoxel extraction. Signal Process Image Commun 33:71–85. https://doi.org/10.1016/j.image.2015.02.005
Tiwari V, Jain SC (2020) Histopathological cells segmentation using exponential grasshopper optimisation algorithm-based fuzzy clustering method. Int J Intell Inf Database Syst 13(2–4):118–138. https://doi.org/10.1504/IJIIDS.2020.109452
Tongbram S, Shimray BA, Singh LS, Dhanachandra N (2021) A novel image segmentation approach using fcm and whale optimization algorithm. J Ambient Intell Humaniz Comput:1–15. https://doi.org/10.1007/s12652-020-02762-w
Use case 1: Nuclei segmentation – andrewjanowczyk (n.d.) http://www.andrewjanowczyk.com/use-case-1-nuclei-segmentation/
Van den Bergh M, Boix X, Roig G, de Capitani B, Van Gool L ((2012, October)) Seeds: Superpixels extracted via energy-driven sampling. In: European conference on computer vision. Springer, Berlin, Heidelberg, pp 13–26. https://doi.org/10.1007/s11263-014-0744-2
Vedaldi A, Soatto S (2008) Quick shift and kernel methods for mode seeking. In: Computer Vision–ECCV 2008: 10th European Conference on Computer Vision, Marseille, France, October 12–18, 2008. Proceedings, Part IV 10 (pp. 705–718). Springer, Berlin Heidelberg. https://doi.org/10.1007/978-3-540-88693-8_52
Veksler O, Boykov Y, Mehrani P (2010) Superpixels and supervoxels in an energy optimization framework. In: European conference on Computer vision. Springer, Berlin, Heidelberg, pp 211–224. https://doi.org/10.1007/978-3-642-15555-0_16
Vishnoi S, Jain AK, Sharma PK (2019) An efficient nuclei segmentation method based on roulette wheel whale optimization and fuzzy clustering. Evol Intel:1–12. https://doi.org/10.1007/s12065-019-00288-5
Wang J, Wang X (2012) VCells: simple and efficient superpixels using edge-weighted centroidal Voronoi tessellations. IEEE Trans Pattern Anal Mach Intell 34(6):1241–1247. https://doi.org/10.1109/TPAMI.2012.47
Wang S, Lu H, Yang F, Yang MH (2011) Superpixel tracking. In: 2011 International Conference on Computer Vision (pp. 1323-1330). IEEE. https://doi.org/10.1109/ICCV.2011.6126385
Wang H, Xiao X, Peng X, Liu Y, Zhao W (2017) Improved image denoising algorithm based on superpixel clustering and sparse representation. Applied Sciences 7(5):436. https://doi.org/10.3390/app7050436
Wei X, Yang Q, Gong Y, Ahuja N, Yang MH (2018) Superpixel hierarchy. IEEE Trans Image Process 27(10):4838–4849. https://doi.org/10.1109/TIP.2018.2836300
Weikersdorfer D, Gossow D, Beetz M (2012) Depth-adaptive superpixels. In: International Conference on Pattern Recognition, pp. 2087–2090
Wolpert DH, Macready WG (1997) No free lunch theorems for optimization. IEEE Trans Evol Comput 1(1):67–82. https://doi.org/10.1109/4235.585893
Wu X, Liu X, Chen Y, Shen J, Zhao W (2018) A graph based superpixel generation algorithm. Appl Intell 48(11):4485–4496. https://doi.org/10.1007/s10489-018-1223-1
Wu C, Zhang L, Zhang H, Yan H (2019) Improved superpixel-based fast fuzzy C-means clustering for image segmentation. In: 2019 IEEE International Conference on Image Processing (ICIP). IEEE, pp 1455–1459. https://doi.org/10.1109/ICIP.2019.8803039
Wu C, Zheng J, Feng Z, Zhang H, Zhang L, Cao J, Yan H (2020) Fuzzy SLIC: fuzzy simple linear iterative clustering. IEEE Trans Circuits Syst Video Technol. https://doi.org/10.1109/TCSVT.2020.3019109
Xiang D, Ban Y, Wang W, Su Y (2017) Adaptive superpixel generation for polarimetric SAR images with local iterative clustering and SIRV model. IEEE Trans Geosci Remote Sens 55(6):3115–3131. https://doi.org/10.1109/TGRS.2017.2662010
Xiang D, Tang T, Quan S, Guan D, Su Y (2019) Adaptive superpixel generation for SAR images with linear feature clustering and edge constraint. IEEE Trans Geosci Remote Sens 57(6):3873–3889. https://doi.org/10.1109/TGRS.2017.2662010
Yang XS (2010) Firefly algorithm, stochastic test functions and design optimisation. Int J Bio-Inspired Comput 2(2):78–84. https://doi.org/10.1504/IJBIC.2010.032124
Yang XS (2010) A new metaheuristic bat-inspired algorithm. Nature Inspired Cooperative Strategies for Optimization (NICSO 2010):65–74. https://doi.org/10.1007/978-3-642-12538-6_6
Yang XS (2012) Flower pollination algorithm for global optimization. In: Unconventional Computation and Natural Computation: 11th International Conference, UCNC 2012, Orléan, France, September 3-7, 2012. Proceedings, vol 11. Springer, Berlin, Heidelberg, pp 240–249. https://doi.org/10.1007/978-3-642-32894-7_27
Yang XS, Deb S (2009) Cuckoo search via Lévy flights. In: Nature & Biologically Inspired Computing, 2009. NaBIC 2009. World Congress on. IEEE, pp 210–214. https://doi.org/10.1109/NABIC.2009.5393690
Yao H, Duan Q, Li D, Wang J (2013) An improved K-means clustering algorithm for fish image segmentation. Math Comput Model 58(3-4):790–798. https://doi.org/10.1016/j.mcm.2012.12.025
Yao J, Boben M, Fidler S, Urtasun R (2015) Real-time coarse-to-fine topologically preserving segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2947–2955
Yuan C, Qin X, Qin Z, Wang R (2018) Image segmentation based on modified superpixel segmentation and spectral clustering. J Eng 2018(16):1704–1711. https://doi.org/10.1049/joe.2018.8320
Zandi M, Mahmoudi-Aznaveh A, Mansouri A (2014) Adaptive matching for copy-move forgery detection. In: 2014 IEEE international workshop on information forensics and security (WIFS). IEEE, pp 119–124. https://doi.org/10.1109/WIFS.2014.7084314
Zhang T, Ramakrishnan R, Livny M (1996) BIRCH: an efficient data clustering method for very large databases. ACM sigmod record 25(2):103–114. https://doi.org/10.1145/235968.233324
Zhang Y, Hartley R, Mashford J, Burn S (2011) Superpixels via pseudo-boolean optimization. In: 2011 International Conference on Computer Vision. IEEE, pp 1387–1394. https://doi.org/10.1109/ICCV.2011.6126393
Zhang Y, Yang C, Wang S, Chen T, Li M, Wang X, He F (2013) LiverAtlas: a unique integrated knowledge database for systems‐level research of liver and hepatic disease. Liver Int 33(8):1239–1248. https://doi.org/10.1111/liv.12173
Zhang X, Thibault G, Decencière E, Marcotegui B, Laÿ B, Danno R, Erginay A (2014) Exudate detection in color retinal images for mass screening of diabetic retinopathy. Med Image Anal 18(7):1026–1043. https://doi.org/10.1016/j.media.2014.05.004
Zhang W, Zhang X, Zhao J, Qiang Y, Tian Q, Tang X (2017) A segmentation method for lung nodule image sequences based on superpixels and density-based spatial clustering of applications with noise. PLoS One 12(9):e0184290. https://doi.org/10.1371/journal.pone.0184290
Zhang Q, Liu Y, Zhu S, Han J (2017) Salient object detection based on super-pixel clustering and unified low-rank representation. Comput Vis Image Underst 161:51–64. https://doi.org/10.1016/j.cviu.2017.04.015
Zhang S, Wang H, Huang W, You Z (2018) Plant diseased leaf segmentation and recognition by fusion of superpixel, K-means and PHOG. Optik 157:866–872. https://doi.org/10.1016/j.ijleo.2017.11.190
Zhong Y, Ma A, Soon Ong Y, Zhu Z, Zhang L (2018) Computational intelligence in optical remote sensing image processing. Appl Soft Comput 64:75–93. https://doi.org/10.1016/j.asoc.2017.11.045
Zhou W, Wu C, Yi Y, Du W (2017) Automatic detection of exudates in digital color fundus images using superpixel multi-feature classification. IEEE Access 5:17077–17088. https://doi.org/10.1109/ACCESS.2017.2740239
Acknowledgements
This work has been partially supported with the grant received in research project under RUSA 2.0 component 8, Govt. of India, New Delhi.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest. The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sasmal, B., Dhal, K.G. A survey on the utilization of Superpixel image for clustering based image segmentation. Multimed Tools Appl 82, 35493–35555 (2023). https://doi.org/10.1007/s11042-023-14861-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-14861-9