Abstract
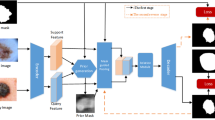
Precise segmentation for skin cancer lesions at different stages is conducive to early detection and further treatment. We propose a weakly supervised semantic segmentation algorithm (CNN-SRR) for dermoscopy images through CNN responding superpixel regions, given that the substantial cost of obtaining perfect pixel annotation for these tasks. CNN-SRR combines a modified classifier based on deep learning and unsupervised superpixel algorithm. The former leverages abundant image-level labeled data to tune parameters to focalize on lesion regions. The extraction of lesion region responses consists of two stages, training a modified CNN classifier and back-propagate peak values of the classifier top layer. Afterward, a test image is over-segmented to a set of primitive superpixels that are merged into a few regions as proposals, several of which are activated as the segmented mask by lesion region responses via non-maximal suppression. Quantified experiments on ISBI2017 and PH2 datasets prove that the proposed algorithm can effectively discriminate lesion regions and the segmentation results even achieve competitive accuracy to the supervised segmentation approaches. We evaluate the proposed CNN-SRR algorithm on ISBI2017 and achieve that the Jaccard coefficient and Accuracy of segmentation task are improved by 12.4% and 3.3% compared with the unsupervised superpixel segmentation algorithm.









Similar content being viewed by others
References
Abbas Q, Celebi ME (2019) Dermodeep-a classification of melanoma-nevus skin lesions using multi-feature fusion of visual features and deep neural network. Multimed Tools Appl, 1–22
Abbas Q, Fondón I, Rashid M (2011) Unsupervised skin lesions border detection via two-dimensional image analysis. Comput Methods Progr Biomed 104 (3):e1–e15
Abuzaghleh O, Barkana BD, Faezipour M (2014) Automated skin lesion analysis based on color and shape geometry feature set for melanoma early detection and prevention. In: IEEE Long Island systems, applications and technology (LISAT) conference 2014, pp 1–6. IEEE
Achanta R, Shaji A, Smith K, Lucchi A, Fua P, Süsstrunk S (2012) Slic superpixels compared to state-of-the-art superpixel methods. IEEE Trans Pattern Anal Mach Intell 34(11):2274–2282
Akram MU, Khan SA (2013) Multilayered thresholding-based blood vessel segmentation for screening of diabetic retinopathy. Eng Comput 29 (2):165–173
Al-Masni MA, Al-antari MA, Choi MT, Han SM, Kim TS (2018) Skin lesion segmentation in dermoscopy images via deep full resolution convolutional networks. Comput Methods Progr Biomed 162:221–231
Argeniano G, Soyer P, De V, Carli P, Delfino M (2002) Interactive atlas of dermoscopy cd
Argenziano G, Fabbrocini G, Carli P, De Giorgi V, Sammarco E, Delfino M (1998) Epiluminescence microscopy for the diagnosis of doubtful melanocytic skin lesions: comparison of the abcd rule of dermatoscopy and a new 7-point checklist based on pattern analysis. Arch Dermatol 134(12):1563–1570
Badrinarayanan V, Kendall A, Cipolla R (2017) Segnet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell 39(12):2481–2495
Balch CM, Buzaid AC, Soong SJ, Atkins MB, Cascinelli N, Coit DG, Fleming ID, Gershenwald JE, Houghton Jr A, Kirkwood JM et al (2001) Final version of the american joint committee on cancer staging system for cutaneous melanoma. J Clin Oncol 19(16):3635–3648
Barata C, Celebi ME, Marques JS (2015) Melanoma detection algorithm based on feature fusion. In: 2015 37th Annual international conference of the IEEE engineering in medicine and biology society (EMBC), pp 2653–2656. IEEE
Bi L, Kim J, Ahn E, Feng D (2017) Automatic skin lesion analysis using large-scale dermoscopy images and deep residual networks. arXiv:1703.04197
Bi L, Kim J, Ahn E, Kumar A, Feng D, Fulham M (2019) Step-wise integration of deep class-specific learning for dermoscopic image segmentation. Pattern Recogn 85:78–89
Celebi ME, Codella N, Halpern A (2019) Dermoscopy image analysis: overview and future directions. IEEE J Biomed Health Inform 23(2):474–478
Celebi ME, Iyatomi H, Schaefer G, Stoecker WV (2009) Lesion border detection in dermoscopy images. Comput Med Imag Graph 33(2):148–153
Celebi ME, Wen Q, Iyatomi H, Shimizu K, Zhou H, Schaefer G (2015) A state-of-the-art survey on lesion border detection in dermoscopy images. Dermoscopy Image Anal 10:97–129
Codella NC, Gutman D, Celebi ME, Helba B, Marchetti MA, Dusza SW, Kalloo A, Liopyris K, Mishra N, Kittler H et al (2018) Skin lesion analysis toward melanoma detection: a challenge at the 2017 international symposium on biomedical imaging (isbi), hosted by the international skin imaging collaboration (isic). In: 2018 IEEE 15th International symposium on biomedical imaging (ISBI 2018), pp 168–172. IEEE
Combalia M, Codella NC, Rotemberg V, Helba B, Vilaplana V, Reiter O, Halpern AC, Puig S, Malvehy J (2019) Bcn20000: dermoscopic lesions in the wild. arXiv:1908.02288
Crandall R Level set implementation. https://github.com/rcrandall/ChanVese/. Accessed 04 Apr 2015
Emre Celebi M, Kingravi HA, Iyatomi H, Alp Aslandogan Y, Stoecker WV, Moss RH, Malters JM, Grichnik JM, Marghoob AA, Rabinovitz HS et al (2008) Border detection in dermoscopy images using statistical region merging. Skin Res Technol 14(3):347–353
Feng X, Yang J, Laine AF, Angelini ED (2017) Discriminative localization in cnns for weakly-supervised segmentation of pulmonary nodules. In: International conference on medical image computing and computer-assisted intervention, pp 568–576. Springer
Freedberg KA, Geller AC, Miller DR, Lew RA, Koh HK (1999) Screening for malignant melanoma: a cost-effectiveness analysis. J Am Acad Dermatol 41(5):738–745
Garcia-Garcia A, Orts-Escolano S, Oprea S, Villena-Martinez V, Garcia-Rodriguez J (2017) A review on deep learning techniques applied to semantic segmentation. arXiv:1704.06857
Gessert N, Sentker T, Madesta F, Schmitz R, Kniep H, Baltruschat I, Werner R, Schlaefer A (2019) Skin lesion classification using cnns with patch-based attention and diagnosis-guided loss weighting. IEEE Transactions on Biomedical Engineering
Gonzalez RC, Woods RE (2006) Digital image processing, 3rd edn. Prentice-Hall, Inc., Upper Saddle River
Greer RO, Marx RE, Said S, Prok LD (2016) Pediatric head and neck pathology. Cambridge University Press. https://doi.org/10.1017/9781316661949
He K, Sun J, Tang X (2010) Guided image filtering. In: European conference on computer vision, pp 1–14. Springer
Jaworek-Korjakowska J (2019) Acral melanocytic lesion segmentation with a convolution neural network (u-net). In: Medical imaging 2019: computer-aided diagnosis, vol 10950, p 109504B. International Society for Optics and Photonics
Karim AM, Güzel MS, Tolun MR, Kaya H, Celebi FV (2019) A new framework using deep auto-encoder and energy spectral density for medical waveform data classification and processing. Biocybern Biomed Eng 39(1):148–159
Karim AM, Kaya H, Güzel MS, Tolun MR, Çelebi FV, Mishra A (2020) A novel framework using deep auto-encoders based linear model for data classification. Sensors 20(21):6378
Kittler H, Pehamberger H, Wolff K, Binder M (2002) Diagnostic accuracy of dermoscopy. Lancet Oncol 3(3):159–165
Li X, Yu L, Chen H, Fu CW, Heng PA (2018) Semi-supervised skin lesion segmentation via transformation consistent self-ensembling model. arXiv:1808.03887
Li Y, Shen L (2018) Skin lesion analysis towards melanoma detection using deep learning network. Sensors 18(2):556
Lissner I, Urban P (2011) Toward a unified color space for perception-based image processing. IEEE Trans Image Process 21(3):1153–1168
Long J, Shelhamer E, Darrell T (2015) Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 3431–3440
Maninis KK, Pont-Tuset J, Arbeláez P, Van Gool L (2016) Convolutional oriented boundaries. In: European conference on computer vision, pp 580–596. Springer
Mendonça T, Ferreira PM, Marques JS, Marcal AR, Rozeira J (2013) Ph 2-a dermoscopic image database for research and benchmarking. In: 2013 35th annual international conference of the IEEE engineering in medicine and biology society (EMBC), pp 5437–5440. IEEE
Oliveira RB, Papa JP, Pereira AS, Tavares JMR (2018) Computational methods for pigmented skin lesion classification in images: review and future trends. Neural Comput Applic 29(3):613–636
Patiño D, Avendaño J, Branch JW (2018) Automatic skin lesion segmentation on dermoscopic images by the means of superpixel merging. In: International conference on medical image computing and computer-assisted intervention, pp 728–736. Springer
Pennisi A, Bloisi DD, Nardi D, Giampetruzzi AR, Mondino C, Facchiano A (2016) Skin lesion image segmentation using delaunay triangulation for melanoma detection. Comput Med Imaging Graph 52:89–103
Schaefer G, Rajab MI, Celebi ME, Iyatomi H (2011) Colour and contrast enhancement for improved skin lesion segmentation. Comput Med Imaging Graph 35(2):99–104
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RG, Barzi A, Jemal A (2017) Colorectal cancer statistics, 2017. CA: a Cancer Journal for Clinicians 67(3):177–193
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv:1409.1556
Soille P (2013) Morphological image analysis: principles and applications. Springer Science & Business Media
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A (2015) Going deeper with convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1–9
Tschandl P, Rosendahl C, Kittler H (2018) The ham10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci Data 5:180161
Tsotsos JK, Culhane SM, Wai WYK, Lai Y, Davis N, Nuflo F (1995) Modeling visual attention via selective tuning. Artif Intell 78(1-2):507–545
Vezhnevets A, Buhmann JM (2010) Towards weakly supervised semantic segmentation by means of multiple instance and multitask learning. In: 2010 IEEE Computer society conference on computer vision and pattern recognition, pp 3249–3256. IEEE
Wei Z, Song H, Chen L, Li Q, Han G (2019) Attention-based denseunet network with adversarial training for skin lesion segmentation. IEEE Access 7:136616–136629
Xie F, Yang J, Liu J, Jiang Z, Zheng Y, Wang Y (2020) Skin lesion segmentation using high-resolution convolutional neural network. Comput Methods Programs Biomed 186:105241
Xie Y, Zhang J, Xia Y, Shen C (2020) A mutual bootstrapping model for automated skin lesion segmentation and classification. IEEE Transactions on Medical Imaging
Yuan Y (2017) Automatic skin lesion segmentation with fully convolutional-deconvolutional networks. arXiv:1703.05165
Zhang J, Bargal SA, Lin Z, Brandt J, Shen X, Sclaroff S (2018) Top-down neural attention by excitation backprop. Int J Comput Vis 126 (10):1084–1102
Zhao Q (2001) Jseg method implementation. cs.joensuu.fi/Zhao/Software/JSEG.zip
Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A (2016) Learning deep features for discriminative localization. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2921–2929
Zhou Y, Zhu Y, Ye Q, Qiu Q, Jiao J (2018) Weakly supervised instance segmentation using class peak response. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 3791–3800
Acknowledgements
This work was partly funded by Natural Science Foundation of China (No.61872225); Introduction and Cultivation Program for Young Creative Talents in Colleges and Universities of Shandong Province (No.173); the Natural Science Foundation of Shandong Province (No.ZR2019ZD04, No.ZR201 5FM010); the Project of Science and technology plan of Shandong higher education institutions Program (No.J15LN20); the Project of Shandong Province Medical and Health Technology Development Program (No.2016WS0577).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interests
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yanfei Hong and Guisheng Zhang contribute equally to this work.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hong, Y., Zhang, G., Wei, B. et al. Weakly supervised semantic segmentation for skin cancer via CNN superpixel region response. Multimed Tools Appl 82, 6829–6847 (2023). https://doi.org/10.1007/s11042-022-13606-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-022-13606-4