Abstract
The development of efficient search engine queries for biomedical images, especially in case of query-mismatch is still defined as an ill-posed problem. Vector-space model is found to be useful for handling the query-mismatch issue. However, vector-space model does not consider the relational details among the keywords and biomedical image search space is not evaluated. Therefore, in this paper, we have proposed a deep learning based fusion vector-space based model. The proposed model enhances the biomedical image query similarity matching approach by fusing the vector space model and convolutional neural networks. Deep learning model is defined by converting the vector-space model to a classification model. Finally, deep learning model is trained to implement the search engine for biomedical images. Extensive experiments reveal that the proposed model achieves significant improvement over the existing models.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Search engines are widely used in various realtime applications. Generally digital libraries and text search engines work in similar fashion. Both utilizes several indexes and utilize words for saving or retrieval of the search space [1, 25, 34]. Various searching and indexing approaches are utilized to implement the search engines. Some popular approaches are as boolean retrieval model, inverted index, etc. [11, 21, 25, 34]. However, the size of indexes become exponentially complex as number of search space increases [25]. Therefore, ranking of indexes is done based upon their retrieval frequency [34]. However, to evaluate the queries where similarity has significantly lesser values, is still a challenging task [10, 14, 31]. To overcome query mismatch issue, query expansion approaches have been implemented to improve the results [28]. Various alternatives or similar query words were utilized to prevent query mismatch issue [24]. Thereafter, linguistic approaches have been implemented such as latent semantic indexing [2], term-document matrix [7], word-net [20], singular value decomposition [8], etc. Latent dirichlet allocation [3, 24, 30]. However, the development of efficient search engine queries especially, in case of query biomedical image-mismatch is still defined as an ill-posed problem.
Figure 1 shows the diagrammatic representation of the biomedical search engine. It clearly shows that an efficient training model is required to build an offline biomedical image search engine. Also, during the online phase users can pass query images and obtained the respective results.
The main contributions of this paper as as:
-
1.
A deep learning based vector-space is proposed for improving the query similarity matching for enhancing the performance of search engines especially for mismatch queries.
-
2.
A softmax function is defined by converting the vector-space model to classification problem.
-
3.
Finally, deep learning model is trained to implement the search engine for biomedical images.
-
4.
Extensive experiments reveal that the proposed model outperforms the competitive models in terms of various performance metrics.
The remaining paper is as: The literature review is discussed in Section 2. Proposed model is mathematically defined in Section 3. Experimental results and discussions are presented in Section 4. Concluding remarks are presented in Section 5.
2 Literature review
Pinho et al. designed a novel biomedical search engine. An extensible model for biomedical images combined with an open-source picture archiving and communication model with profile-based capabilities has been utilized [22]. Long designed a novel search engine model for a supplemental health applications. Supplemental federated search engine was also designed. Performance was evaluated on federated search engine along with website usability testing results [19]. Faroo has reviewed many biomedical search engines and found that the development of biomedical search engine is still an open area of research [6].
Hochberg et al. designed biomedical search engine to diagnose diabetes. Different models such as decision tree, logistic regression, linear regression, and random forest to diagnose diabetic patients [12]. Ye et al. designed COVID-19-related query logs to develop search engines. It was significant to learn about the epidemic’s influence on users’ search behavior and improve search engine to tackle comparable pandemic outbreaks in the future [32]. Young et al. implemented a search engine for diagnose HIV infected patients. A negative binomial approach was designed to estimate HIV infected patients by considering a subgroup of predictor keywords recognized by lasso regression. The Google search data was integrated with existing HIV reports [33]
Fagroud et al. designed a novel internet of things (IoT) search engine. With the advancement in IoT networks and the enhancement of the number of IoT resources, searching the data of IoT, learning IoT, recognize and list of the associated resources have become a necessity, which became possible with the presence of a various kind of IoT search engines. [5]. Kopanos et al. implemented an VarSome i.e., human genomic variant search engine [17]. Doulani et al. discussed a Scopus database and google scholar search engine. The statistical population of 118 researchers who were active in social- scientific network from 29 governmental universities were utilized. t-test and pearson correlation coefficient were implemented for search engine analysis [4].
Recently many researchers have designed various machine learning models such as to classify various kind of applications [9, 13]. However, the majority of the existing models suffer from the over-fitting issue [15, 16, 29]. Therefore, in this paper, a novel fusion mdoel by using DCNN and vector space model is proposed to achieve better results.
3 Proposed model
In this section, initially, vector-space model is discussed. Thereafter, deep convolutional neural network (DCNN) is presented. Finally, DCNN based vector-space model is discussed.
3.1 Vector-space model
In this paper, we have focused on the vector-space based query similarity matching approach for improving the performance of search engines especially for biomedical image-mismatch queries.
The vector-space model defines search space and queries as group of vector indexes. Weights define the significance of biomedical image features in query Q and image space D [18] as:
To defines weights for biomedical image search space vector, lq − aDq [26] is utilized. In lq − aDq [26], weights are computed using two factors lqak i.e., frequency of word k in Da and occurrence of k in collected search space (Dqk). Dqk requires weight scaling.
W defines total search space in Dqk. Inverse biomedical image search space frequency (aDqk) of k is defined as:
It augments aDq. However, it may convert aDq frequent terms with low degree [25]. A composite weight is defined by integrating Dqk and aDqk. Therefore, in lQ − aDq weighting, the weight of k ∈ Da is represented as:
It provides significantly more weights to words having higher frequency [23, 27]. By considering lq − aDq, the vector-space model computes cosine similarity (cos 𝜃) among biomedical image search space and query vectors [23, 27]. cos 𝜃 defines vector details of Da and Q, respectively, by utilizing the dot multiplication of two vectors and also the multiplication of their respective Euclidean values. cos 𝜃 can be evaluated as:
By using (5), dot multiplication \(|\overrightarrow {D_{a}}\parallel \overrightarrow {Q}|\) can be evaluated as \({\sum }_{k-1}^{U} M_{Qk}\times M_{ak}\). Here, MQ,k shows weight of k in query q. U defines size of word. \(|\overrightarrow {D_{a}}\parallel \overrightarrow {Q}|\) defines the multiplication of Euclidean values and can be evaluated as \({\sum }_{k-1}^{U} M^{2}_{Q,k} {\sum }_{k-1}^{U} {M^{2}_{a}}k\). Integration of these variables define the similarity among Da and Q as:
Equation (6) predicts the normalization normalization impact in a search engine. However, vector-space model does not consider the relational details among the keywords and biomedical image search space is not evaluated.
3.2 Deep learning model
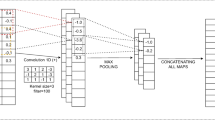
In this section, deep convolutional neural network (DCNN) based vector-space model is defined. Our goal is to predict such a combination of Q and D which can provide more accurate results. DCNN requires various convolution filters to squeeze local features (please see Fig. 2).
Consider there is single channel which can be defined as:
where \( C \in {\mathbb {R}^{n \times k}} \). n shows the size of input biomedical image. k represents the enclosed dimension of every input factor. In convolution process, a filter \( {\textbf {m}} \in \mathbb {R}^{lk} \) is required in implementing to successive l biomedical images to bring potential features as:
Here, ci:i+l− 1 is the integration of ci,...,ci+l− 1. \( b \in \mathbb {R} \) is a bias. f represents a non-liner activation function like relu. Thereafter, filter m move towards \( \left \{ {{{\textbf {c}}_{1:l}},{{\textbf {c}}_{2:l + 1}},...,{{\textbf {c}}_{n - l + 1:n}}} \right \} \), then following feature map can be obtained:
Thereafter, max-pool is implemented on x to obtain the maximum value \( \hat x = {\max \limits } \{ {\textbf {x}}\} \). It defines the final feature extracted by m. It obtains the dominated feature set of every filter. CNN computes various feature sets by using numerous filters with different sizes. The obtained feature sets contain a vector as
Here, s defines the number of filters. The softmax (sf) layer is then used to compute the estimated probability distribution as:
Consider a training data (xi, yi) in which \( {y^{i}} \in \left \{ {1,2, {\cdots } ,c} \right \} \) defines matched image query for search engine of xi and approximated probability of DCNN is \( \tilde {y_{j}^{i}} \in [0,1] \) for every label \( j \in \left \{ {1,2, \cdots ,c} \right \} \). The estimated error can be computed as:
where c shows the number of labels of xi. \( if\{ \dot \} \) define as an indicator and if{yi = j} = 1 if yi = j, if{yi = j} = 0 otherwise. The stochastic gradient descent is employed to update the DCNN attributes and adopt Adam optimizer.
4 Performance analysis
The proposed model is applied to the benchmark search engine dataset. The comparison of the proposed technique is drawn with the state-of-art models such as Decision tree, Logistic regression, Support vector machine, Artificial neural network, Random forest, Naive Bayes, k nearest neighbour (k-NN), Adaboost, SVM-Random forest, CNN, and Gradient boosting. The experiments are performed on core i7 3.80 GHz, 32-GB RAM, and 15M cache on MATLAB 2019a software.
Figure 3 shows the validation, training and testing analysis of proposed model. It is found that the proposed model converges at very fast speed during the training process. At 262nd epoch, the proposed model achieves the best training and validation results, respectively. Thus, the proposed model obtains significantly lesser binary-cross entropy values, i.e., loss during the model building process.
To evaluate the performance of the proposed model, median and degree of uncertainty values (i.e., median ± IQR × 1.5) are evaluated by repeating the experiments 50 times. We have used 65% dataset for training, 15% for validation, and 20% for testing, respectively. The fraction of training is set to be 65% because the obtained dataset is small in size. Other experiments are also considered by changing the fractions of training data. But it is found that the significant performance is found when the fraction of the training data is 65%.
To draw comparisons among the proposed and the existing models, confusion matrix-based measures are used. These measures are accuracy, specificity, sensitivity, area under curve (AUC) and f-measure.
Tables 1 and 2 depict the training and testing analysis of the proposed model for biomedical search engine dataset. Various confusion matrix based metrics like accuracy, sensitivity, specificity, f-measure, and AUC are used to compute the effectiveness of the proposed model over the existing models. From these tables, it is found that the proposed automated model provides significantly better results as compared to the existing model. As the proposed model achieves significantly better sensitivity and specificity values, therefore, a fast and efficient search engine similarity algorithm is proposed.
Table 3 shows web image search engines analysis among the proposed and the existing models. It is found that the proposed model outperforms the competitive web image serach engines.
5 Conclusion
From the extensive review, it has been found that the vector-space model did not consider the relational details among the biomedical contents and image search space. Therefore, a fused DCNN and vector-space based biomedical image query similarity matching approach was proposed for improving the performance of biomedical search engines. DCNN model was defined by converting the vector-space model to classification problem. Finally, biomedical image search engine was trained. Extensive experiments have been drawn by using the proposed and the competitive models for search engines. The proposed model has shown significant improvement over the existing biomedical search engines.
References
Basavegowda HS, Dagnew G (2020) Deep learning approach for microarray cancer data classification. CAAI Trans Intell Technol 5(1):22–33
Bigelow JL, Edwards A, Edwards L (2016) Detecting cyberbullying using latent semantic indexing. In: Proceedings of the first international workshop on computational methods for CyberSafety, pp 11–14
Cimiano P, Schultz A, Sizov S, Sorg P, Staab S (2009) Explicit versus latent concept models for cross-language information retrieval. In: IJCAI, vol 9, pp 1513–1518
Doulani A, Shabani Z, Baradar R (2020) Information science academic members of iranian public universities sharing information resources in researchgate social scientific network: It’s relation on their scientific output in scopus database and google scholar search engine. J Payavard Salamat 14(1):53–64
Fagroud FZ, Ben Lahmar EH, Amine M, Toumi H, El Filali S (2019) What does mean search engine for iot or iot search engine. In: Proceedings of the 4th international conference on big data and internet of things, pp 1–7
Faroo D (2017) Search engine optimization for medical publishing, Reconstruct Rev 7 (4)
Furnas GW, Deerwester S, Durnais ST, Landauer TK, Harshman RA, Streeter LA, Lochbaum KE (2017) Information retrieval using a singular value decomposition model of latent semantic structure. In: ACM SIGIR Forum, vol 51. ACM, New York, pp 90–105
Gao W, Guo Y, Wang K (2016) Ontology algorithm using singular value decomposition and applied in multidisciplinary. Clust Comput 19 (4):2201–2210
Ghosh S, Shivakumara P, Roy P, Pal U, Lu T (2020) Graphology based handwritten character analysis for human behaviour identification. CAAI Trans Intell Technol 5(1):55–65
Gupta A, Singh D, Kaur M (2020) An efficient image encryption using non-dominated sorting genetic algorithm-iii based 4-d chaotic maps. J Ambient Intell Humaniz Comput 11(3):1309–1324
Gupta B, Tiwari M, Lamba SS (2019) Visibility improvement and mass segmentation of mammogram images using quantile separated histogram equalisation with local contrast enhancement. CAAI Trans Intell Technol 4(2):73–79
Hochberg I, Daoud D, Shehadeh N, Yom-Tov E (2019) Can internet search engine queries be used to diagnose diabetes? analysis of archival search data. Acta Diabetol 56(10):1149–1154
Kaur M, Singh D, Kumar V (2020) Color image encryption using minimax differential evolution-based 7d hyper-chaotic map. Appl Phys B 126(9):1–19
Kaur M, Singh D, Kumar V, Sun K (2020) Color image dehazing using gradient channel prior and guided l0 filter. Inf Sci 521:326–342
Kaur M, Singh D, Sun K, Rawat U (2020) Color image encryption using non-dominated sorting genetic algorithm with local chaotic search based 5d chaotic map. Futur Gener Comput Syst 107:333–350
Kaur M, Singh D, Uppal RS (2019) Parallel strength pareto evolutionary algorithm-ii based image encryption. IET Image Process 14(6):1015–1026
Kopanos C, Tsiolkas V, Kouris A, Chapple CE, Aguilera MA, Meyer R, Massouras A (2019) Varsome: The human genomic variant search engine. Bioinformatics 35(11):1978
Lee DL, Chuang H, Seamons K (1997) Document ranking and the vector-space model. IEEE Softw 14(2):67–75
Long BA (2017) Addressing a discovery tool’s shortcomings with a supplemental health sciences-specific federated search engine. J Electron Res Med Lib 14(3-4):101–113
Miller GA (1995) Wordnet: A lexical database for english. Commun ACM 38(11):39–41
Osterland S, Weber J (2019) Analytical analysis of single-stage pressure relief valves. Int J Hydromechatron 2(1):32–53
Pinho E, Godinho T, Valente F, Costa C (2017) A multimodal search engine for medical imaging studies. J Digit Imag 30(1):39–48
Ross NC, Wolfram D (2000) End user searching on the internet: An analysis of term pair topics submitted to the excite search engine. J Am Soc Inf Sci 51(10):949–958
Schütze H, Hull DA, Pedersen JO (1995) A comparison of classifiers and document representations for the routing problem. In: Proceedings of the 18th annual international ACM SIGIR conference on research and development in information retrieval, pp 229–237
Schütze H, Manning CD, Raghavan P (2008) Introduction to information retrieval, vol 39. Cambridge University Press, Cambridge
Singhal A, et al. (2001) Modern information retrieval: A brief overview. IEEE Data Eng Bull 24(4):35–43
Spink A, Wolfram D, Jansen MB, Saracevic T (2001) Searching the web: The public and their queries. J Amer Soc Inform Sci Technol 52(3):226–234
Voorhees EM (1994) Query expansion using lexical-semantic relations. In: SIGIR’94. Springer, pp 61–69
Wang R, Yu H, Wang G, Zhang G, Wang W (2019) Study on the dynamic and static characteristics of gas static thrust bearing with micro-hole restrictors. Int J Hydromechatron 2(3):189–202
Wei X, Croft WB (2006) Lda-based document models for ad-hoc retrieval. In: Proceedings of the 29th annual international ACM SIGIR conference on research and development in information retrieval, pp 178–185
Wiens T (2019) Engine speed reduction for hydraulic machinery using predictive algorithms. Int J Hydromechatron 2(1):16–31
Ye Z, Mao J, Liu Y, Zhang M, Ma S (2020) Investigating covid-19-related query logs of chinese search engine users. Proc Assoc Inform Sci Technol 57(1):e424
Young SD, Zhang Q (2018) Using search engine big data for predicting new hiv diagnoses. PloS one 13(7):e0199527
Zobel J, Moffat A (2006) Inverted files for text search engines. In: ACM Comput Surv (CSUR), vol 38, pp 6–es
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mishra, R., Tripathi, S.P. Deep learning based search engine for biomedical images using convolutional neural networks. Multimed Tools Appl 80, 15057–15065 (2021). https://doi.org/10.1007/s11042-020-10391-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-020-10391-w