Abstract
Several methods utilizing common spatial pattern (CSP) algorithm have been presented for improving the identification of imagery movement patterns for brain computer interface applications. The present study focuses on improving a CSP-based algorithm for detecting the motor imagery movement patterns. A discriminative filter bank of CSP method using a discriminative sensitive learning vector quantization (DFBCSP-DSLVQ) system is implemented. Four algorithms are then combined to form three methods for improving the efficiency of the DFBCSP-DSLVQ method, namely the kernel linear discriminant analysis (KLDA), the kernel principal component analysis (KPCA), the soft margin support vector machine (SSVM) classifier and the generalized radial bases functions (GRBF) kernel. The GRBF is used as a kernel for the KLDA, the KPCA feature selection algorithms and the SSVM classifier. In addition, three types of classifiers, namely K-nearest neighbor (K-NN), neural network (NN) and traditional support vector machine (SVM), are employed to evaluate the efficiency of the classifiers. Results show that the best algorithm is the combination of the DFBCSP-DSLVQ method using the SSVM classifier with GRBF kernel (SSVM-GRBF), in which the best average accuracy, attained are 92.70% and 83.21%, respectively. Results of the Repeated Measures ANOVA shows the statistically significant dominance of this method at p < 0.05. The presented algorithms are then compared with the base algorithm of this study i.e. the DFBCSP-DSLVQ with the SVM-RBF classifier. It is concluded that the algorithms, which are based on the SSVM-GRBF classifier and the KLDA with the SSVM-GRBF classifiers give sufficient accuracy and reliable results.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
In recent years, a number of different strategies have been investigated for the control of robotic limbs such as hand or foot. For instance,the electromyogram (EMG) and the electroencephalogram (EEG) biosignals or a combination of both have been used for limb control. This research focuses on the analysis of the EEG signals to estimate motor imagery movement patterns. When actual movements happen in the EEG signals, the event-related desynchronization (ERD) patterns occurs firstly, and then the event-related synchronization (ERS) potential patterns immediately appear in the EEG signal. Various methods using common spatial patterns (CSP) have been developed to detect motor imagery movement patterns in the EEG signals automatically. The CSP is a linear map that maximizes the difference of variances between two classes. The CSP procedure was first introduced by Ramoser et al. [29] for extracting discriminative features of imaginary hand movements by spatial filters and a linear classifier. The application of the CSP is then extended to detect foot imagery movements; for example, Wang et al. [43] employed the CSP to distinguish the hand and foot imaginary movement patterns. As the CSP method is sensitive to noise [25], various strategies have been investigated to control and reduce the noise before the CSP calculation. Lemm et al. [20] proposed the common spatio-spectral pattern approach to reduce the noise by a time delay embedding method. The time delay embedding method is able to select and adjust the best frequency filter bands for individual electrodes. Dornhege et al. [5] developed the CSP further by optimizing a spectral and spatial filter to improve differentiation rates among multichannel trials; the approach was designated as the common sparse spectral spatial pattern (CSSSP) method. A further method reducing noise is the sub-band common spatial pattern (SBCSP) method developed by Novi et al. [25]. In the SBCSP method, the EEG signal is decomposed into sub-frequencies by the Gabor filter bank. A CSP algorithm is then used on individually filtered sub-frequency bands. The sub-frequency bands are subsequently scored and removed by recursive band elimination method. Finally, a support vector machine (SVM) classifier is utilized to classify the CSP features. Ang et al. [2] developed the FBCSP algorithm and compared with the SBCSP. In the FBCSP method, first a bandpass filter bank is used and, then the CSP approach is applied. Two feature selection algorithms, named as the mutual information best individual feature (MIBIF) and Mutual Information based rough set reduction, are employed to select the most informative pairs of the frequency bands and CSP features. Ang et al. [1] extended the FBCSP by extending the two-class CSP to a four-class method using the Divide-and-Conquer, Pair-Wise, and One-Versus-Rest methods. Thomas et al. [42] developed the discriminative filter bank before the CSP (DFBCSP) method to improve the FBCSP results. In the DFBCSP, a set of nine frequency ranges are employed as a filter bank. The CSP is then applied to extract features. The features are then selected and classified by a mutual information approach and the SVM classifier, respectively. Sun et al. [37] developed a CSP-based method, namely the sliding window discriminative CSP (SWDCSP). The sliding window in the SWDCSP shifts over the range of frequency bands to organize a bandpass filter bank. The CSP is then applied and features are selected using the affinity propagation method. The selected features are finally classified by the SVM method. Jamaloo et al. [16] combined the filter bank common spatial pattern (FBCSP) and DFBCSP methods. Results were improved by utilizing the discrimination sensitive learning vector quantization (DSLVQ) with the DFBCSP (DFBCSP-DSLVQ) to assign weights to the CSP features for discriminant feature scattering. Xu et al. [45] improved the Regularized CSP (RCSP) approach [21] using a transfer learning and cosine similarities methods. For improvement, two RCSP algorithms with a cosine measurements are utilized. The cosine functions are replaced with covariance measurements for estimating the subject’s differences. The cosine computes the similarities of spatial filters among the target subjects and improves the RCSP method results. In the study of Zou et al. [48], a combination of the CSP, the broad learning (BL) and neural network (NN) are employed for multi-classification. With the CSP approach, brain patterns are extracted from the EEG signal, while the BL learning and the NN methods are used for feature extraction and classifications. Another recent study is performed by Khalaf et al. [17] utilizing a combined of the CSP and wavelets for feature extraction to measure the EEG and cerebral blood velocity within motor imagery task.
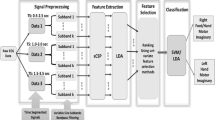
The KPCA and KLDA algorithms have been successfully used for feature selection in image processing and signal processing for mapping features into higher feature space dimension for feature selection and classification [14, 22, 32, 46]. Hoffmann et al. [14] used the KPCA algorithm for the breast-cancer cytology and handwritten detection based on the image processing. The KPCA algorithm was found to be more successful than other performed methods for cancer detection. Widjaja et al. [44] successfully used the KPCA approach to extract respiratory signals from an ECG for sleep apnea. Zhao et al. [47] used seven types of feature extraction to extract features and classify female facial expressions. The PCA, LDA, KPCA and KLDA methods are considered as the best ones among the seven considerable methods. Of the four methods listed, the KLDA algorithm has previously been found to give the best results with the lowest dimension. In a classification study based on the BCI, Hoffmann et al. [14] showed that the SVM with a RBF kernel has a greater impact on the classification accuracy than other kernels such as polynomial, linear and Sigmoid. In our previous studies [9, 11,12,13, 24], the GRBF kernel has higher accuracy and robustness. The procedure of this study is: recording nine EEG data based on the BCI competition III, dataset IVa (the designed task is the same as the BCI Competition III dataset IVa task); implementing the existed DFBCSP-DSLVQ method; applying a frequency sub-band selection method to find the best sub-bands; using the KLDA and KPCA algorithms, in which the kernel is GRBF, to select the features; and finally utilizing different combinations to classify the selected features, namely the SVM with the radial basis function (RBF) kernel (SVM-RBF), SVM with the GRBF kernel (SVM-GRBF), soft margin support vector machine (SSVM) with the RBF kernel (SSVM-RBF), SSVM with the GRBF kernel (SSVM-GRBF), the NN, and the K-nearest neighbor (K-NN). The obtained results are then compared with DFBCSP-DSLVQ method with the SVM-RBF classifier. The diagram of the procedure is presented in Fig. 1. The structure of the rest of the paper is presented as follows: Section 2 explains the implemented methods individually, such as filter bank, CSP, sub-band selection, DSLVQ, KLDA, KPCA, GRBF, SSVM, NN, and K-NN. Section 3 explains the experimental setup that includes: the number of subjects, task regulations and the procedure of trials. Section 4 includes the obtained results in terms of accuracy, robustness, paired T-test and Repeated Measures ANOVA with the Post-hoc using Tukey correction evaluations. Section 5 discusses the obtained results. Finally, Section 6, is the conclusion from the discussion and obtained results.
2 Method
The base of the present motor imagery algorithm is the CSP procedure that searches and finds the best projection direction in the feature space to minimize the variance of features in one class and maximize it in the other class [25, 42].
2.1 Filter bank
A filter bank is used to extract six sub-band frequencies from the EEG signal (8-12Hz, 12-16Hz, 16-20Hz, 20-24Hz, 24-28Hz, and 28-32Hz) to enable the CSP procedure to extract imagery movement features [2].
2.2 CSP calculation
In the CSP procedure, the average of normalized EEG covariance for each trial is computed [16]:
where uc is the covariance matrix, which is normalized and averaged in class c; while the number of trials for each class is denote by m. The covariance matrices of the classes are combined as shown in (2) to compute the superposition of the eigenvalue to whiten the covariance matrix of each class in (3).
where Z0 is the eigenvectors matrix, P is the diagonal eigenvalue matrix and T is the transpose operator. The whitening transformation matrix is (3):
where B is the whitened matrix and λ is a diagonal eigenvalue matrix of U. The whitening matrix is applied on each covariance class by (4) and sorted in decreasing order:
where Fc is the new eigenvector, in which the smallest eigenvalue is assigned to one class and the largest eigenvalue is assigned to the other class. In order to consider the correctness of calculations, the sum of the eigenvalues of the two classes must be unity (5), (6):
where, ϕ1 and ϕ2 are the diagonal matrix of eigenvalues for the both classes and I is identity matrix. Mapping of each movement trial is computed by applying the CSP values (\({A}_{CSP}={{Z}^{T}}W\)) on the EEG signal (X(i)) for each trial. The CSP for a new signal is obtained by (7):
Based on the previous study [16], only the first and last rows of the Q are used for extracting variance features (Wi = var(Qi)). In order to extract the variance features, a procedure of frequency sub-band selection is applied and explained in the following section:
2.3 Frequency sub-band selection
The filter bank decomposes the EEG signal into six sub-frequency bands. The most informative frequency sub-bands are distinguished by two differential assessing algorithms. First, the difference between the CSP features across all trails of the imagery right hand and foot movement classes is computed as follows:
where i is the number of the variance features (W ), \(W_{Hk}^{1}\) and \(W_{Hk}^{l}\) are the first and last rows of each feature from the imagination of right hand and foot movement classes in the f frequency band, respectively; and, k is the index of the selected frequency band. The maximum average of the difference is selected as the informative frequency band. The second differential assessment is the difference between the first and last rows of the CSP features for the six sub-bands. The sub-bands, which has the largest difference, are selected for further computations. The extracted variance features form the selected sub-bands are then weighted by a DSLVQ approach to have more distinct values for classification. The DSLVQ method is presented in the following section:
2.4 Discriminative sensitive learning vector quantization
The learning vector quantization (LVQ) is an effective approach to classify single trial patterns [18, 26]. The goal of the LVQ algorithm is to distribute samples in the multi-dimension feature space [3, 28]. The DSLVQ method computes and assigns weights to the features to indicate their importance. Three improvements are employed to improve the generalization of the LVQ: I- weights are calculated by an iterative training algorithm for each dimension based on distances; II- a vector of weights is produced for each feature in (9); and III- the iterative training is used to optimize and select one scalar weight for each feature in (10). The selected optimal weights estimate distances for the iterative learning (t) procedure [26]:
where W(t) is the present feature vector and W(t + 1) the shift from present to the new feature vector, respectively. Wnew(t) is the new weight vector calculated in (10), and t is the number of the iteration. β(t) is the learning rate to find the optimum weight; if it is smaller than one, a decision will then be made between the new and current weights in the specific iteration. The new weight Wnew(t) is calculated based on the Euclidean distance between the present and new weight as follows:
where distO is the distance among training data in the individual classes in the feature space, and distC is the distance of features between classes. For better projection between classes, weighted features are mapped into a higher feature space with the KPCA or KLDA approaches for better separability, which is presented in the following two parts.
2.5 Kernel principal component analysis
The KPCA is a method introduced by Schölkopf et al. [33] to solve the principal component analysis (PCA) problem. The PCA is an approach for image processing [33], image compression [27] and bio-signal processing [44]. The PCA method is based on an assumption that only a few principal components are needed to reconstruct the dataset. The principal components are computed by orthogonal mapping from the input space to the feature space and called as factors or latent variables. The PCA is used to extract only those features from the input training data that contains higher amounts of information [35]. Factors are obtained by calculating the orthogonal transformations from the input data in the coordinate system. The PCA is calculated in the following three steps: I- calculation of the covariance matrix by (11); II- Computing the projection of the test data on the largest eigenvectors by (18); and III- calculating eigenvector in the feature space through (11) and (12) as follows [33, 35]:
where n is the number of observations. The observations are centered by means of \(\sum \limits _{i=1}^{n}{{{W}_{j}}=0}\).
where λ is the eigenvalue for the KPCA, λ ≥ 0 and V is the eigenvector for the KPCA. In some applications, the extracted features do not have linear relation to the input data. In order to solve the linearity problem, the PCA is extended with the KPCA. The KPCA algorithm uses a nonlinear map (φ) to transfer the training data into a higher dimension (\(\varphi ({{x}_{j}}):{{R}^{n}}\to S\)) to extract the informative components [44]. In this study, the nonlinear map is the GRBF kernel, as described in part 3.8. The PCA limitation is solved by an integral operator kernel function technique. The procedure uses the covariance (\(\overline {Cov}\)) in (13) to compute the eigenvalues and eigenvectors related to the KPCA:
where V is the eigenvector m by m dimensions.
where α is the normalizing coefficient eigenvector that must satisfy the criterion \({{\lambda }_{n}}({{\alpha _{i=1}}^{n}}.{{\alpha _{j=1}}^{n}})=1\):
H is a n × n matrix. The (14), (15), and (16) are combined to compute the eigenvalues:
Finally, the projection of the sample data φ(xj) on the eigenvector vn in the computed feature space is obtained as follows:
The second method is the KLDA with the same purpose of better projection to have better data separation by increasing the feature space dimension.
2.6 Kernel linear discriminant analysis (KLDA)
Similar to the PCA, the LDA is for dimension reduction for classification. In order to solve the linearity limitation of the LDA, Roth et al. and Hastie et al. [8, 31] extended the algorithm by introducing kernel functions. The basic concept of the LDA is to project observations of c classes in c − 1 dimensions using a maximizing criterion. Maximizing the ratio between Sb and Sw variables that are assigned to between-group scattering and within-group scattering, respectively [30]. In the projection procedure, the eigenvalue of \( S_{w}^{-1}{{S}_{b}}\) is calculated and sorted in decreasing order. The first value is the largest value of the eigenvalues that contain the most informative data. In the LDA, the observations are centered (\(\sum \limits _{i=1}^{n}{{{W}_{j}}=0}\)), and then the Sb and Sw matrices are defined [31]:
where nc is the number of the pattern and \(W_{i}^{(c)}\) is the observation parameter. In the second step, the mapping matrix (ψ) is formed to maximize the ratio between the Sb and Sw variables (21):
The optimum result for V is the eigenvalues related to the KLDA, which are computed by Sbvi = λiSwvi. In order to extend the LDA to KLDA as a nonlinear map (Γ), which is constructed by nonlinear mapping. In this paper, the nonlinear map is the GRBF that is defined in part 3.8. The KLDA is then applied on the feature space (Γ : Rn → S, w →Γ(w)) and the selected features are centered \((\sum \limits _{i=1}^{n}{{{W}_{i}}=0})\). The constructed feature space is finally classified by the SSVM and SVM classifiers with the RBF and GRBF kernels, the NN and K-NN classifiers in the following parts:
2.7 Classifiers
The extracted and selected variance features are then classified with different classifiers, namely SVM-RBF, SVM-GRBF, SSVM-GRBF, K-NN, NN [6, 10, 11, 15, 24, 34, 36, 40].
2.8 Generalized radial basis function
For the classification, the SSVM classifier with the GRBF kernel is explained as follows: to overcome the RBF’s limitation, the generalized Gaussian distribution function was proposed based on the Gaussian function [6]. The GRBF approach is more flexible and stable than the RBF and can be applied to behaviors with wider variety and less data. The generalized Gaussian function is presented in (22):
where w > 0, τ > 0, and center are the width, shape and center of the distribution, respectively. γ is a factorial extension function defined as:
The width of distribution, w, defined as:
where σ is the standard deviation. The τ parameter in (22) is employed to generalize the RBF that should be adjusted. By increasing τ (\(\tau \to \infty \)), the distribution shape will become the uniform distribution; by decreasing (τ → 0), the shape will become the impulse distribution; and for the two special cases τ = 1 and τ = 2, the density shape will be the Laplacian and Gaussian distribution, respectively (Fig. 2). The generalized Gaussian distribution function fits better the shape of the kernel with the distributed data (Fig. 3).
Different τ values for the generalized Gaussian distribution (center = 2, w = 1), (22)
Based on the previous studies [10, 14] , the integration of the SVM classifier with the GRBF kernel is a powerful method for classification. Regularizing the SVM is highly impressive for controlling the cost function than the traditional SVM [4]. The GRBF is used as a kernel of the SSVM and SVM that computations are presented in the following parts.
2.9 Soft margin SVM
The SSVM is a modified SVM classifier [4]. In the SSVM a training set (\({{W}_{i}}\in {{R}^{n}},i=1,...,l\)) with two classes (c = [1,2]) is utilized. In the basic SVM, a linear hyperplane in the feature space is defined (\({{W}^{T}}{{x}_{i}}+b=0\)) as a decision boundary with two conditions in (25):
where W is the feature weights, and b is the bias. In the basic SVM, the challenge is finding the maximum margin distance between the classes by employing some features that are called support vectors (SVs). The decision function based on the SVs is:
where x is the input of the decision function test data. In the nonlinear cases, a nonlinear map φ(xi) is employed to separate the features (W ) in the high dimension feature space. However, the modified hyperplane has cost in each training iteration. The cost is optimized by a regularization parameter in (27) [41]:
where R is a positive regularization parameter, ζi is the cost function for the ith training set, and ζ(W;x, Y ) ≡ (1 − YWTx)2. By significantly increasing the dimension of W, the problem of duality appears and is solved by the Lagrange’s theorem in (28):
where e is a l × 1 identity matrix, α is the Lagrange coefficient, k is the kernel function, \(K({{x}_{i}},{{x}_{j}})\equiv \varphi {{({{x}_{i}})}^{T}}\varphi ({{x}_{j}})\), and the optimal value of Y is obtained by \(W=\sum \limits _{i=1}^{l}{{{Y}_{i}}{{\alpha }_{i}}\varphi ({{x}_{i}})}\). Finally, the decision boundary is altered as follows:
2.10 K-nearest neighbor
The K-NN is a supervised robust classifier, and is based on the Euclidean distance. The K-NN algorithm is trained by the closest samples in the feature space and tested by votes based on the distance that are assigned to a new data. K is the number of neighbors that are selected, for instance, K = 5 means five closest neighbors to the new data in the feature space. If the test data has more than three votes to a specific class, the test data is assigned to that class. In this study, different nearest neighbors are selected experimentally, and the best result was obtained by the 1-nearest neighbor classifier. In this experiment, the dataset is divided into 10 folds. For each training, one fold is selected for testing and the rest are used for training.
2.11 Neural network (NN)
The NN is a well-known supervised classifier and is used in various fields and applications. In this study, the implemented NN configuration has 210 input features for training (75% of data), 15 features for cross validation and 55 for testing. The configuration of the NN is 210 inputs; two hidden layers that include 10 and 15 neurons respectively and employed the Sigmoid transfer functions; one linear neuron in the output layer. The NN is trained by back propagation training technique. The number of hidden layers and neurons are achieved experimentally.
3 Experiment setup
The EEG data of five subjects (aa, al, av, aw, ay) from the BCI Competition III, Dataset IVa, were used. The EEG data of every subjects contain 280 trials of right hand (140 trial) and foot imagery movements (140 trial), which are recorded by 118 channels in the EEG recorder. The sampling frequency was 100 Hz. In order to extract the CSP features, a window of two-second width i.e., from 0.5 to 2.5 seconds after the cue stimulation, was selected. The stimulation cue was a picture to instigate the subject to imagine the hand or foot movement. Despite the BCI III Dataset IVa EEG data, nine more EEG data sets were recorded (9 subjects S1 to S9, the average age of subjects is 27 years old). Ethics approval was obtained from the Research Ethics Committee (REC) of Lappeenranta University of Technology. Subjects signed a letter of satisfaction and they were informed of the experiment before attending it. The designed task for recording the EEG is depicted in Fig. 4. The task is presented as follows: I- a black page is showed for 500 msec; II- a cross sign is presented at the center to attract the attention of subjects; III- send a marker on the EEG data and show the pictures for imaginary or real movements for 1 sec; IV- a black screen is displayed to imagine a movement; and V- rest for 3.5 to 4 secs. The cycle is displayed for 280 times individually. The utilized portable device is the ENOBIO32, which is suitable for BCI applications. The Enobio32 has 32 channels to record the EEG signal. The sampling frequency was set at 500 Hz, which is down sampled to 100 Hz.
4 Results
Utilizing the sub-band selection approach, the 8-12Hz and 12-16 Hz ranges were found as the most informative frequency ranges. Results based on the DFBCSP-DSLVQ method with 12 combinations of the KLDA, KPCA, RBF and GRBF algorithms are presented in Tables 1, 2, 3 and 4. The NN and K-NN classifiers are used for comparison with the 12 combinations. In order to show the most informative frequencies of the selected sub-bands and channels, spectrograms using the short-time fourier transform (STFT) and Welch power spectrum estimates for channel C3 and channel O1 (the most and the least related channels) are used. In the STFT and Welch power, three frequency sub-bands, 8-12Hz, 12-16 Hz and 16-20 Hz, are depicted in Figs. 5 and 6. The KLDA and KPCA approaches are utilized to map the features of the changed projection of coordinates into higher dimension feature space and to enable better scattering for classification with lower feature space dimension. Different combinations of feature selection algorithms and classifiers are implemented to evaluate performance of the algorithms. The results are evaluated with the accuracy and robustness, and presented in Tables 1 to 4. Because the EEG dataset IVa of the BCI competition III has 118 channels and our EEG data has 32 channels, Tables 1 and 2 are formed separately to present the results. Tables 3 and 4 are presented for comparing the robustness and SVs for the two dataset. A statistical paired T-test is applied on the obtained features of the hand and foot imagination movements detection for individuals, and results are presented in the discussion part. The paired T-test gives information on the meaningfulness of the within-group feature changes. Robustness is defined as the variation of accuracies in one method (\((100-({\max \limits } (accuracy)-{\min \limits } (accuracy))\)). Finally, the best accuracy is determined using the Repeated Measures ANOVA statistical analysis [7] with the Post-hoc using Tukey correction test [38]. Different combinations of the SVM-based classifiers select different numbers of SVs and these numbers play an important role in selecting decision boundary. The average number of SVs are presented in Tables 3 and 4. In addition, the scatter plot of the features and selected SVs are showed in Figs. 7, 8, 9, 10, 11, 12, 13, 14, 15, 16 and 17.
Welch power spectral estimation for imaginary hand movement in channel C3 in frequency bands of 8-12 Hz, 12-16 Hz and 16-20 Hz for subject aa. The maximum amplitude of the three frequency bands is considered. For channel C3, the highest power intensities for the frequency bands of 8-12 Hz, 12-16 Hz and 16-20 Hz frequency bands are -7.44 dB/Hz and -17.68 dB/Hz and -20.33 dB/Hz, respectively. The spectrum used the STFT method for imaginary hand movement in the channel C3 for a 2-sec window in the three main informative frequency ranges
Welch power spectral estimation for imaginary hand movement incChannel O1 in frequency bands of 8-12 Hz, 12-16 Hz and 16-20 Hz for subject aa. The maximum amplitude of the three frequency bands is considered. For the channel O1 the highest power intensities for the frequency bands of 8-12 Hz, 12-16 Hz and 16-20 Hz are -38.89 dB/Hz, -41.15 dB/Hz and -44.09 dB/Hz respectively. The spectrum used the STFT method for imaginary hand movement imagination in the channel O1 for a 2-sec window in the three main informative frequency ranges
5 Discussion
One successful method for extracting the imagery movement patterns for controlling an artificial limb based on the EEG biosignals is the CSP. The aim of the CSP algorithm is to search for the best projection direction in the feature space and to minimizes the variance for one class and maximizes for the other class [23]. One recent method is the DFBCSP-DSLVQ approach, on which this study is based. The aim of this study is to improve the DFBCSP-DSLVQ method for the BCI applications. 14 different combinations of algorithms are employed to improve the DFBCSP-DSLVQ method with the SVM-RBF classifier [16], namely the KLDA, KPCA, RBF, GRBF, SVM, SSVM, K-NN, and NN. In order to find the most informative frequency sub-bands, the EEG signals are divided into six frequency bands. The CSP with sub-band selection algorithms are applied to select the most informative sub-bands. Then, the sub-bands, 8-12 Hz and 12-16 Hz, were chosen for feature classification. The best channels and frequency sub-bands for detecting the imagery movement patterns were estimated by power spectrum (STFT) and power spectral estimate (Welch) methods, as shown in Figs. 5 and 6. When there is not enough information about data, one effective approach for finding the best channel and frequency range to compute the power spectral density estimate by the Welch method. The Welch method shows the power of signal as frequency and returns the estimation of a one-sided power spectrum density. The one-sided power spectrum density contains the total signal power in half of the Nyquist interval [19]. In the Welch procedure, signal is divided into eight segments and each segment is multiplied by the Hamming window. Channels C3 and O1 were found as the most and the least informative channels, respectively for extracting the imagery right hand movement features. In the case of the aa subject, the most informative frequencies for the hand movement trials were found in the first two sub-bands, which are 8-12 Hz and 12-16 Hz (Figs. 5 and 6). Informative information can be concluded from the amplitude of the frequency bands, when the frequencies reach the maximum values. The maximum power intensities during a 2 secs window for the imagery of right hand movement in the first two sub-bands are -8.44 and -17.68 dB/Hz for channel C3 and -38.89 and the minimum intensity power are -41.15 dB/Hz for channel O1, (Figs. 5 and 6). The spectrogram in Fig. 5 visually shows the maximum power of the EEG signals for the right hand after cue stimulation. In the spectrograms, the observable discontinuous green and blue colors are the frequencies masked with the band pass filter. The brighter colors are interpreted as areas that are more informative. In Figs. 5 and 6, it is observable that power of channel C3 is higher and the spectrogram in that area is brighter. The most informative frequency bands are revealed by two differential assessment methods [16]. In order to find the best sub-bands mathematically, two differential assessments are explained as in (8) in part 2.3. After applying the DFBCSP, the axes projection of the feature space is changed. The DFBCSP produces high discrepancy between the two classes by maximizing the variance of features for one class and minimizing for the second class. As shown in Figs. 7 to 17, the features scattering of the imaginary for right hand movement class is separated from the imaginary foot movement class. Hence, the differential values between the minimum and maximum variances increased. The sub-bands with the largest differential values are selected as the aim frequency band [16]. The most informative frequency bands for the subjects were determined as 8-12 Hz and 12-16 Hz. Therefore, the two frequency bands are employed instead of the six frequency bands.
Scattering of the first row (Feature 1) versus the last row (Feature 2) of the DFBCSP-DSLVQ features for subject aa. 98 SVs are selected for the SVM with the RBF (SVM-RBF) classifier (the base method case 4 in Table 1)
Next, the variance features are obtained from the selected sub-bands (8-12 Hz and 12-16 Hz) and weighted by the DSLVQ, which is a modified LVQ method showing the effectiveness of the features individually. The DSLVQ weights are based on the Euclidean distance and showed as positions of the features in the feature space and the number of times, at which each feature is repeated. Weights are then calculated by a weight-learning iteration procedure for each dimension in (9). The weighted values have direct relation with the number of repetitions; the more number of repetitions the smaller the weight. Therefore, the algorithm uses a very small coefficient to reduce the impact of the EEG background (repetitive values) in the calculations. On the other hand, the single trial waves such as the ERDs have different values against the EEG background. Hence, the DSLVQ algorithm generates a greater coefficient value. Additionally, when the features in the training set are classified correctly, the related weights will increase and vice versa. Thus, the effect of the EEG background signal decreases and the effect of the single trial movements increases. Finally, the imaginary hand and foot movement features are well distinguished by the computed weights, and this leads to high accuracies with low variation (robustness). Tables 3 and 4 contain the robustness values for subjects individually. The elapsed time for the DSLVQ processing was 0.050688 seconds that is suggested for the real-time applications.
The KLDA and KPCA algorithms are employed to achieve better data separation by selecting features. The KLDA maximize between-group scattering over within-group scattering, \({\max \limits } (\frac {\det (Sb)}{\det (Sw)})\), whereas the KPCA procedure seeks a space to maximize the variance direction in the feature space. The KPCA is not sensitive to the number of training dataset [23]. The KPCA- and KLDA-based algorithms have disadvantages of missing the spatial information for classification. In other words, the KPCA causes removal of some projection directions that reject low variance values [10]. The selected kernel for the KLDA and KPCA is the GRBF. In the KLDA and KPCA procedure, the GRBF increase the feature space dimension to separate the data of the classes by the generalized Gaussian function, which is updated automatically for individual subjects.
The results are presented in Tables 1 to 4. Tables 1 and 3 include results based on the EEG data with 118 channels (the BCI competition III, dataset IVa). Tables 2 and 4 include results based on the EEG data with 32 channels that are recorded in our Laboratory. In order to classify the obtained features from the selected channels and sub-bands, the following algorithms are utilized: the SVM-RBF, SSVM-RBF, SVM-GRBF, SSVM-GRBF, K-NN, and NN. The basic classifier method of this paper is the DFBCSP-DSLVQ with the SVM-RBF kernel (case4). Tables 1 and 3 show that the NN classifier has 13.42% lower accuracy and 25.87% higher robustness in comparison to the case 4. The cause for the low accuracy is the insufficient number of data for training the NN. The next classifier is the K-NN, which is applied on the computed features. This approach is based on the Euclidean distance between the test data and the specified (k − th) neighbors around it. Different neighbors were tested experimentally and the 1-NN algorithm was chosen as the best neighbor. Tables 1 and 3 show 1.35% higher accuracy and 7.14% lower robustness in comparison with the case 4, respectively, whilst Tables 2 and 4 shows 1.70% and 23.70% higher accuracy and robustness, respectively. The next classifiers for imaginary pattern recognition are the SVM-based methods. For improving the basic method, different combinations of the SVM, SSVM, RBF and GRBF algorithms are employed. For instance, the GRBF kernel is used as a kernel of the SVM that needs optimization for the Gaussianity. The parameter’s Gaussianity determines the precise shape of the Gaussian function for the distribution (Figs. 2 and 3).
The cases 1 to 12 are divided into three groups, A (cases 1 to 4), B (cases 5 to 8) and C (9 to 12). The two best methods are compared with the case 4 [16]. Group A results in Tables 1 and 3 show that the case 1 is the best improvement of the case 4 with higher accuracy and robustness of 12.00% and 23.21%, respectively. Also, Tables 2 and 4 show the significant increases of 15.92% and 17.06% for accuracy and robustness, respectively. The results of group A in Tables 2 and 4, reveal that the SSVM classifier with the GRBF kernel has the most significant impact on the results. Figures 7 to 10 show the functionality of different classifier combinations on the data scattering of group A. The best classier is the one that achieves the best accuracy with less number of selected SVs. Figure 10 depicts that the SSVM-GRBF combination with 33 SVs, reaches the average accuracy of 92,70% with robustness of 83.21% in group A (Tables 1 and 3). Group B is the combination of group A with the KLDA feature selection algorithm and the best result is the case 5. The average accuracies in Tables 1 and 3 show the increases of 4.66% and 16.06% in accuracy and robustness, respectively, when compared with the case 4. Results in Tables 2 and 4 show that the case 5 attained higher accuracy and robustness of 9.74% and 20.66%, respectively, when compared with the case 4. Different combinations of the classifiers and kernels are used in the group B. Results in Tables 2 and 4 show that the KLDA with the SSVM-GRBF has the best impacts on the data. Figure 11 shows that the number of selected SVs is 47, which is less than the number of selected SVs in Fig. 7 (case 4). Regarding Tables 1 and 3, the best average of accuracy and robustness in group B are 85.36% and 76.07%, receptively. Group C is the combination of group A with the KPCA feature selection algorithm. Tables 1 and 3 show that the result in the group C is achieved in the case 11 with 75.78% average accuracy and 60.36% robustness, which are lower than case 4. Tables 2 and 4 show that the case 9 is the best method with 60.99% average accuracy and 56.79% robustness in group C.
In consideration among groups, Tables 1 and 2 are used to analyze the impact of the algorithms on the data classification. Tables 1 and 2 show the effectiveness of the KLDA and KPCA algorithms with classifiers in three groups. Classifiers with and without the KLDA or KPCA from different groups are selected for comparison such as cases 1, 5 and 9 from groups of A, B and C, respectively. Table 1 shows that the methods using the classifiers without the KPCA or KLDA feature selection algorithms achieved higher accuracy than the methods with using the feature selection algorithms. This claim is observed in four following categories: cases 1, 5, 9; cases 2, 6, and 10; cases 4, 8 and 12; and cases 3, 7 and 11. The same results are observed in Table 2, except two categories of cases 2, 6 and 10, and cases 4, 8 and 12 that shows higher accuracy for the methods with the KLDA algorithm. The KLDA improves the accuracy results in comparison with the methods with KPCA algorithm. By comparing the scattering of subject 5 (S5) (Figs. 13 to 15) and the subject aa (Figs. 16 and 17), it is revealed that, the KLDA algorithm does not increase the discrepancy for some features of subjects such as aa, al, aw, etc. But the KLDA increase the feature’s separation such as S5. Therefore, experimentally we can recognize the efficiency of the KLDA or KPCA. Tables 1 and 3 show that Case 1 (group A) has the best average accuracy and robustness of 91.27% and 83.21%, respectively. From Tables 2 and 4, the Case 1 has the best average accuracy and robustness of 78.03% and 73.66%, respectively.
In order to find the best method, statistical analysis are employed. Based on the data distribution, normal or non-normal, parametric or non-parametric statistical methods such as paired T-test and Wilcoxon Signed Rank [39] are used, respectively. Because, before feature extraction the data is normalized between zero and one, the paired T-test is utilized for considering the meaningfulness of the features for individual subjects to figure out if the extracted features are informative for classification. Then, the Repeated Measures ANOVA test with the Post-hoc using Tukey correction [38] test algorithm is applied on the accuracies to find the best algorithm. The paired T-test results showed that in total 14 subject’s features were changed significantly (T-test, p < 0.05) between the imaginary hand and foot movements. In addition, based on the Repeated Measures ANOVA with the Post-hoc using Tukey correction test the DFBCSP-DSLVQ method with the SSVM-GRBF classifier significantly had the best result, Table 1 with 92.07% (ANOVA, p < 0.05) and Table 2 with 78.03% (ANOVA, p < 0.05). The segregation of the classes is considered by scattering plots of the DFBCSP-DSLVQ features. Therefore, three scatter plots for the imaginary hand and foot movements are depicted in Figs. 13 to 15. Figures 13 and 15 show that the aggregation of the computed features based on the DFBCSP-DSLVQ algorithm and DFBCSP-DSLVQ method with the KPCA is more than the DFBCSP-DSLVQ method with the KLDA algorithm. The DFBCSP-DSLVQ with the KLDA feature selection method has better effect on the feature’s discrepancy, as shown in Fig. 14. The results in Tables 1 and 2 show that the classification of the DFBCSP-DSLVQ features with the SSVM-GRB combination produce better accuracies and robustness than the other combinations. The reason of better result for the SSVM-GRBF method without the KLDA is removing some projection directions that reject low variance values, although increased the discrepancy. The order of classifiers from the best to the worst classifier performance is as follows: the SSVM-GRBF, SVM-GRBF, SSVM-RBF SVM-RBF, NN, and K-NN.
6 Conclusion
In this study, 14 approaches are implemented with the aim of improving the DFBCSP-DSLVQ with the SVM-RBF method for recognizing the imagery hand and foot movement patterns. In the processing, the informative frequency sub-bands are selected, variance features are extracted, and then weighted by the DSLVQ method. The KLDA and KPCA algorithms are then utilized to map the features into a new feature space for feature selection and better data separation. For classification, different combination of algorithms are then utilized, namely the NN, K-NN, SVM and SSVM with both RBF and GRBF kernels. Results indicate that the SSVM-GRBF classifier brings out the best average accuracy and robustness of 92.70% and 83.21% with ANOVA p < 0.05, respectively.
Abbreviations
- EEG::
-
electroencephalogram
- BCI::
-
brain computer interface
- CSP::
-
common spatial pattern
- ERD::
-
event related desynchronization
- LDA::
-
linear discriminant analysis
- KLDA::
-
kernel linear discriminant analysis
- PCA::
-
principal component analysis
- KPCA::
-
kernel principal component analysis
- LVQ::
-
learning vector quantization
- DSLVQ::
-
discriminative sensitive learning vector quantization
- SSVM::
-
soft margin support vector machine
- GRBF::
-
generalized radial basis function
- RCSP::
-
Regularized CSP
- BL::
-
board learning
- SV::
-
support vector
- K-NN::
-
K-nearest neighbor
- NN::
-
neural network
- Min::
-
minimum
- Max::
-
maximum
- STFT::
-
short-time Fourier transform
- DFBCSP-DSLVQ::
-
discriminative filter bank common spatial pattern method using discriminative sensitive learning vector quantization
- DFBCSP-DSLVQ-SVM-RBF::
-
DFBCSP method using DSLVQ for weighting and SVM classifier with RBF kernel
- DFBCSP-DSLVQ-KLDA-SSVM-GRBF::
-
DFBCSP method using DSLVQ for weighting, followed by KLDA and SSVM classifier with GRBF kernel
- DFBCSP-DSLVQ-KPCA-SSVM-GRBF::
-
DFBCSP method using DSLVQ for weighting, followed by KPCA and SSVM classifier with GRBF kernel
References
Ang KK, Chin ZY, Wang C, Guan C, Zhang H (2012) Filter bank common spatial pattern algorithm on bci competition iv datasets 2a and 2b. Frontiers in Neuroscience, pp 6–39
Ang KK, Chin ZY, Zhang H, Guan C (2008) Filter bank common spatial pattern (FBCSP) in brain-computer interface. In: 2008 ieee International Joint Conference on Neural Networks (IEEE World Congress on Computational Intelligence), IEEE, pp 2390–2397
Blankertz B, Muller KR, Curio G, Vaughan TM, Schalk G, Wolpaw JR, Schlogl A, Neuper C, Pfurtscheller G, Hinterberger T et al (2004) The bci competition 2003: progress and perspectives in detection and discrimination of eeg single trials. IEEE Transactions on Biomedical Engineering 51(6):1044–1051
Chang CC, Lin CJ (2011) Libsvm: a library for support vector machines. ACM Transactions on Intelligent Systems and Technology (TIST) 2(3):27
Dornhege G, Blankertz B, Krauledat M, Losch F, Curio G, Muller KR (2006) Combined optimization of spatial and temporal filters for improving brain-computer interfacing. IEEE Transactions on Biomedical Engineering 53(11):2274–2281
Fernandez-Navarro F, Hervas-Martinez C, Sanchez-Monedero J, Gutierrez PA (2011) Melm-grbf: a modified version of the extreme learning machine for generalized radial basis function neural networks. Neurocomputing 74(16):2502–2510
Girden ER (1992) ANOVA: Repeated measures (No. 84). Sage
Hastie T, Tibshirani R, Buja A (1994) Flexible discriminant analysis by optimal scoring. Journal of the American statistical association 89(428):1255–1270
Hekmatmanesh A, Jamaloo F, Wu H, Handroos H, Kilpeläinen A Common spatial pattern combined with kernel linear discriminate and generalized radial basis function for motor imagery-based brain computer interface applications. In: AIP Conference Proceedings 2018 Apr 27, vol 1956, no 1, AIP Publishing, p 020003
Hekmatmanesh A, Mikaeili M, Sadeghniiat-Haghighi K, Wu H, Handroos H, Martinek R, Nazeran H (2017) Sleep spindle detection and prediction using a mixture of time series and chaotic features. Advances in Electrical and Electronic Engineering 15(3):435
Hekmatmanesh A, Noori SMR, Mikaili M (2014) Sleep spindle detection using modified extreme learning ma- chine generalized radial basis function method. In: 2014 22nd Iranian Conference on Electrical Engineering (ICEE), IEEE, pp 1898–1902
Hekmatmanesh A, Wu H, Li M, Nasrabadi AM, Handroos H (2019) Optimized mother wavelet in a combination of wavelet packet with detrended fluctuation analysis for controlling a remote vehicle with imagery movement: a brain computer interface study. In: New Trends in Medical and Service Robotics. Springer, Cham, pp 186–195
Hekmatmanesh A, Wu H, Motie-Nasrabadi A, Li M, Handroos H (2019) Combination of discrete wavelet packet transform with detrended fluctuation analysis using customized mother wavelet with the aim of an imagery-motor control interface for an exoskeleton. Multimedia Tools and Applications, pp 1–20
Hoffmann H (2007) Kernel pca for novelty detection. Pattern Recogn 40(3):863–874
Hsu CW, Chang CC, Lin CJ A practical guide to support vector classification
Jamaloo F, Mikaeili M (2015) Discriminative common spatial pattern sub-bands weighting based on distinction sensitive learning vector quantization method in motor imagery based brain-computer interface. Journal of Medical Signals and Sensors 5(3):156
Khalaf A, Sejdic E, Akcakaya M Common spatial pattern and wavelet decomposition for motor imagery EEG-fTCD brain-computer interface. Journal of Neuroscience Methods, 2019
Kohonen T (1990) The self-organizing map. proceedings of the ieee, 78
Krauss TP, Shure L, Little JN (1994) Signal processing toolbox for use with Matlab
Lemm S, Blankertz B, Curio G, Muller KR (2005) Spatio-spectral filters for improving the classification of single trial eeg. IEEE Transactions on Biomedical Engineering 52(9):1541–1548
Lotte F, Guan C (2011) Regularizing common spatial patterns to improve BCI designs: unified theory and new algorithms. IEEE Transactions on biomedical Engineering 58(2):355–62
Mahato S, Paul S (2019) Electroencephalogram (EEG) signal analysis for diagnosis of major depressive disorder (MDD): a review. In: Nanoelectronics, Circuits and Communication Systems. Springer, Singapore, pp 323–335
Martınez AM, Kak AC (2001) Pca versus lda. IEEE transactions on pattern analysis and machine intelligence 23(2):228–233
Noori SMR, Hekmatmanesh A, Mikaeili M, Sadeghniiat-Haghighi K (2014) K-complex identification in sleep eeg using melm-grbf classifier. In: 2014 21th Iranian Conference on Biomedical Engineering (ICBME), IEEE, pp 119–123
Novi Q, Guan C, Dat TH, Xue P (2007) Sub-band common spatial pattern (sbcsp) for brain-computer interface. In: 2007. CNE’07. 3rd International IEEE/EMBS Conference on Neural Engineer- ing, IEEE, pp 204–207
Ozgen C (2010) Realization of a cue based motor imagery brain computer interface with its potential application to a wheelchair. PhD thesis, MIDDLE EAST TECHNICAL UNIVERSITY
Park S, Lee JJ, Yun CB, Inman DJ (2008) Electro-mechanical impedance-based wireless structural health monitoring using pca- data compression and k-means clustering algorithms. J Intell Mater Syst Struct 19(4):509–520
Pregenzer M, Pfurtscheller G (1995) Distinction sensitive learning vector quantization (dslvq)-application as a classifier based feature selection method for a brain computer interface, pp 433–436
Ramoser H, Muller- Gerking J, Pfurtscheller G (2000) Optimal spatial filtering of single trial eeg during imagined hand movement. IEEE Transactions on Rehabilitation Engineering 8(4):441–446
Ro D, Pe H (1973) Pattern classification and scene analysis
Roth V, Steinhage V (2000) Nonlinear discriminant analysis using kernel functions. In: Advances in Neural Information Processing Systems, pp 568–574
Santamaria L, James C (2018) Using brain connectivity metrics from synchrostates to perform motor imagery classification in EEG-based BCI systems. Healthcare Technology Letters 5(3):88–93
Scholkopf B, Smola A, Muller KR (1998) Nonlinear component analysis as a kernel eigenvalue problem. Neural Computation 10(5):1299–1319
Scholkopf B, Smola AJ (2001) Learning with kernels: support vector machines, regularization, optimization, and beyond. MIT press
Scholkopf B, Smola AJ (2002) Learning with kernels: support vector machines, regularization, optimization, and beyond. MIT press
Subasi A, Ercelebi E (2005) Classification of eeg signals using neural network and logistic regression. Computer methods and programs in biomedicine 78(2):87–99
Sun G, Hu J, Wu G (2010) A novel frequency band selection method for common spatial pattern in motor imagery based brain computer interface. The 2010 International Joint Conference on Neural Networks (IJCNN), IEEE, pp 1-6
Surhone LM, Timpledon MT, Marseken SF Tukey’s Range Test. (VDM Publishing, 2010). at, https://books.google.fi/books?id=b_OycQAACAAJ&dq=Tukey+statistical+test&hl=en&sa=X&redir_esc=y
Surhone LM, Timpledon MT, Marseken SF Wilcoxon Signed-Rank Test. at, https://books.google.com/books?id=gclXYgEACAAJ&dq=Wilcoxon+statistical+test&hl=en&sa=X&ved=0ahUKEwid_NLH6IndAhVLEawKHVNjCCsQ6wEIMjAB (VDM Publishing, 2010)
Suykens JA, Vandewalle J (1999) Least squares support vector machine classifiers. Neural Processing Letters 9(3):293–300
Suykens JA, Vandewalle J (1999) Least squares support vector machine classifiers. Neural processing letters 9(3):293–300
Thomas KP, Guan C, Lau CT, Vinod AP, Ang KK (2009) A new discriminative common spatial pattern method for motor imagery brain–computer interfaces. IEEE Trans Biomed Eng 56(11):2730–2733
Wang Y, Gao S, Gao X (2006) Common spatial pattern method for channel selection in motor imagery based brain-computer interface. In: IEEE-EMBS 2005. 27th Annual International Conference of the Engineering in Medicine and Biology Society, 2005, IEEE, pp 5392–5395
Widjaja D, Varon C, Dorado A, Suykens JA, Van Huffel S (2012) Application of kernel principal compo- nent analysis for single-lead-ecg-derived respiration. IEEE Trans Biomed Eng 59(4):1169–1176
Xu Y, Wei Q, Zhang H, Hu R, Liu J, Hua J, Guo F (2018) Transfer learning based on regularized common spatial patterns using cosine similarities of spatial filters for Motor-Imagery BCI. Journal of Circuits, Systems and Computers 5:1950123
Ye B, Qiu T, Bai X, Liu P (2018) Research on recognition method of driving fatigue state based on sample entropy and kernel principal component analysis. Entropy 20(9):701
Zhao X, Zhang S (2012) Facial expression recognition using local binary patterns and discriminant kernel locally linear embedding. EURASIP journal on Advances in signal processing, pp 1–20
Zou J, She Q, Gao F, Meng M (2018) Multi-task motor imagery eeg classification using broad learning and common spatial pattern. In: International Conference on Intelligence Science. Springer, Cham, pp 3–10
Funding
Open access funding provided by LUT University.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hekmatmanesh, A., Wu, H., Jamaloo, F. et al. A combination of CSP-based method with soft margin SVM classifier and generalized RBF kernel for imagery-based brain computer interface applications. Multimed Tools Appl 79, 17521–17549 (2020). https://doi.org/10.1007/s11042-020-08675-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-020-08675-2