Abstract
Background
Grain length, width, weight, and the number of grains per panicle are crucial determinants contributing to yield in cereal crops. Understanding the genetic basis of grain-related traits has been the main research object in crop science.
Methods and results
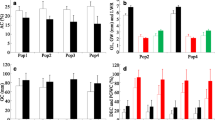
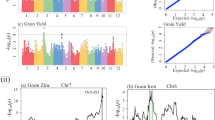
Kerala has a collection of different rice landraces. Characterization of these valuable genetic resources for 39 distinct agro-morphological traits was carried out in two seasons from 2017 to 2019 directly in farmers field. Most characteristics were polymorphic except ligule shape, leaf angle, and panicle axis. The results of principal component analysis implied that leaf length, plant height, culm length, flag leaf length, and grain-related traits were the principal discriminatory characteristics of rice landraces. For identifying the genetic basis of key grain traits of rice, three multi locus GWAS models were performed based on 1,47,994 SNPs in 73 rice accessions. As a result, 48 quantitative trait nucleotides (QTNs) were identified to be associated with these traits. After characterization of their function and expression, 15 significant candidate genes involved in regulating grain width, number of grains per panicle, and yield were identified.
Conclusions
The detected QTNs and candidate genes in this study could be further used for marker-assisted high-quality breeding of rice.



Similar content being viewed by others
Data Availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Code Availability
Not applicable.
References
Ray DK, Mueller ND, West PC, Foley JA (2013) Yield trends are insufficient to double global crop production by 2050. PLoS ONE 8:e66428. https://doi.org/10.1371/journal.pone.0066428
Lu Y, Chuan M, Wang H et al (2022) Genetic and molecular factors in determining grain number per panicle of rice. Front Plant Sci 13. https://doi.org/10.3389/fpls.2022.964246
Li R, Li M, Ashraf U et al (2019) Exploring the relationships between yield and yield-related traits for rice varieties released in China from 1978 to 2017. Front Plant Sci 10:543. https://doi.org/10.3389/fpls.2019.00543
Bhandari A, Sandhu N, Bartholome J et al (2020) Genome-wide association study for yield and yield related traits under reproductive stage drought in a diverse indica-aus Rice Panel. Rice (N Y) 13:53. https://doi.org/10.1186/s12284-020-00406-3
Ponce K, Zhang Y, Guo L et al (2020) Genome-wide association study of grain size traits in Indica Rice Multiparent Advanced Generation Intercross (MAGIC) Population. Front Plant Sci 11:395. https://doi.org/10.3389/fpls.2020.00395
Zhang G, Wang R, Ma J et al (2021) Genome-wide association studies of yield-related traits in high-latitude japonica rice. BMC Genom Data 22:39. https://doi.org/10.1186/s12863-021-00995-y
Wang N, Chen H, Qian Y et al (2023) Genome-wide association study of rice grain shape and chalkiness in a worldwide collection of Xian accessions. Plants 12:419. https://doi.org/10.3390/plants12030419
Semwal D, Pradheep K, John KJ et al (2019) Status of Rice (Oryza sativa L.) Genepool Collected from Western Ghats Region of India: gap analysis and diversity distribution mapping using GIS tools. Ind Jour Plant Gene Resour 32:166. https://doi.org/10.5958/0976-1926.2019.00021.4
Beena R, Kirubakaran S, Nithya N et al (2021) Association mapping of drought tolerance and agronomic traits in rice (Oryza sativa L.) landraces. BMC Plant Biol 21:484. https://doi.org/10.1186/s12870-021-03272-3
Peringottillam M, Kunhiraman Vasumathy S, Selvakumar HKK, Alagu M (2022) Genetic diversity and population structure of rice (Oryza sativa L.) landraces from Kerala, India analyzed through genotyping-by-sequencing. Mol Genet Genomics 297:169–182. https://doi.org/10.1007/s00438-021-01844-4
IRRI (2013) Standard evaluation system for Rice (SES). International Rice Research Institute (IRRI), pp 1–65
IBM Corp (2011) IBM SPSS Statistics for Windows Version 20.0
Wei T, Simko V (2017) R Package “Corrplot”: Visualization of a Correlation Matrix (Version 0.84)
Rosseel Y (2012) lavaan: an R Package for Structural equation modeling. J Stat Softw 48:1–36. https://doi.org/10.18637/jss.v048.i02
Epskamp S (2015) semPlot: unified visualizations of structural equation models. Struct Equation Modeling: Multidisciplinary J 22:474–483. https://doi.org/10.1080/10705511.2014.937847
R Core Team (2018) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue
Poland JA, Rife TW (2012) Genotyping-by‐Sequencing for Plant Breeding and Genetics. The Plant Genome 5:-Wiley Online Library. https://acsess.onlinelibrary.wiley.com/doi/full/https://doi.org/10.3835/plantgenome2012.05.0005
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Zheng X, Levine D, Shen J et al (2012) A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28:3326–3328. https://doi.org/10.1093/bioinformatics/bts606
Zhang Y-W, Tamba CL, Wen Y-J et al (2020) mrMLM v4.0.2: an R platform for multi-locus genome-wide Association Studies. Genomics Proteom Bioinf 18:481–487. https://doi.org/10.1016/j.gpb.2020.06.006
Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265. https://doi.org/10.1093/bioinformatics/bth457
Volante A, Desiderio F, Tondelli A et al (2017) Genome-wide analysis of japonica Rice Performance under Limited Water and permanent flooding conditions. Front Plant Sci 8:1862. https://doi.org/10.3389/fpls.2017.01862
Volante A, Tondelli A, Aragona M et al (2017) Identification of bakanae disease resistance loci in japonica rice through genome wide association study. Rice 10:29. https://doi.org/10.1186/s12284-017-0168-z
Supek F, Bošnjak M, Škunca N, Šmuc T (2011) REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6:e21800. https://doi.org/10.1371/journal.pone.0021800
Manjunatha GA, Elsy CR, Rajendran P et al (2018) Agro-morphological characterization of rice (Oryza sativa L.) landraces of Wayanad, Kerala. J Pharmacogn Phytochem 7:1409–1414
Kovinich N, Kayanja G, Chanoca A et al (2015) Abiotic stresses induce different localizations of anthocyanins in Arabidopsis. Plant Signal Behav 10:e1027850. https://doi.org/10.1080/15592324.2015.1027850
Roy S, Rathi RS, Misra AK et al (2014) Phenotypic characterization of indigenous rice (Oryza sativa L.) germplasm collected from the state of Nagaland, India. Plant Genet Resour 12:58–66. https://doi.org/10.1017/S1479262113000282
Saha SR, Hassan L, Haque MA et al (2019) Genetic variability, heritability, correlation and path analyses of yield components in traditional rice (Oryza sativa L.) landraces: variability and traits association in rice. J Bangladesh Agricultural Univ 17:26–32
Bassuony NN, Zsembeli J (2021) Inheritance of some flag leaf and yield characteristics by half-diallel analysis in rice crops (Oryza sativa L). Cereal Res Commun 49:503–510. https://doi.org/10.1007/s42976-020-00115-z
Zhong H, Liu S, Sun T et al (2021) Multi-locus genome-wide association studies for five yield-related traits in rice. BMC Plant Biol 21:364. https://doi.org/10.1186/s12870-021-03146-8
Mogga M, Sibiya J, Shimelis H et al (2019) Correction: diversity analysis and genome-wide association studies of grain shape and eating quality traits in rice (Oryza sativa L.) using DArT markers. PLoS ONE 14:e0212078. https://doi.org/10.1371/journal.pone.0212078
Rohilla M, Singh N, Mazumder A et al (2020) Genome-wide association studies using 50 K rice genic SNP chip unveil genetic architecture for anaerobic germination of deep-water rice population of Assam, India. Mol Genet Genomics 295:1211–1226. https://doi.org/10.1007/s00438-020-01690-w
Soumya PR, Burridge AJ, Singh N et al (2021) Population structure and genome-wide association studies in bread wheat for phosphorus efficiency traits using 35 K Wheat Breeder’s Affymetrix array. Sci Rep 11:7601. https://doi.org/10.1038/s41598-021-87182-2
Kirkby RA (1986) Uses of on-farm trials in a crop improvement program. In: Pigeonpea Workshop, 8–11 April. p 10PP
Nguyen DT, Gomez LD, Harper A et al (2020) Association mapping identifies quantitative trait loci (QTL) for digestibility in rice straw. Biotechnol Biofuels 13:165. https://doi.org/10.1186/s13068-020-01807-8
Zhang Y-M, Jia Z, Dunwell JM (2019) Editorial: the applications of New Multi-Locus GWAS Methodologies in the genetic dissection of Complex Traits. Front Plant Sci 10:100. https://doi.org/10.3389/fpls.2019.00100
Huang N, Parco A, Mew T et al (1997) RFLP mapping of isozymes, RAPD and QTLs for grain shape, brown planthopper resistance in a doubled haploid rice population. Mol Breeding 3:105–113. https://doi.org/10.1023/A:1009683603862
Luo LJ, Li ZK, Mei HW et al (2001) Overdominant epistatic loci are the primary genetic basis of inbreeding depression and heterosis in rice. II. Grain yield components. Genetics 158:1755–1771
Hua JP, Xing YZ, Xu CG et al (2002) Genetic dissection of an elite rice hybrid revealed that heterozygotes are not always advantageous for performance. Genetics 162:1885–1895. https://doi.org/10.1093/genetics/162.4.1885
Yoshida S, Ikegami M, Kuze J et al (2002) QTL analysis for Plant and Grain characters of sake-brewing Rice using a doubled Haploid Population. Breed Sci 52:309–317. https://doi.org/10.1270/jsbbs.52.309
Thomson MJ, Tai TH, McClung AM et al (2003) Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor Appl Genet 107:479–493. https://doi.org/10.1007/s00122-003-1270-8
Sun L, Li X, Fu Y et al (2013) GS6, a member of the GRAS gene family, negatively regulates grain size in rice. J Integr Plant Biol 55:938–949. https://doi.org/10.1111/jipb.12062
Wang J, Yu H, Xiong G et al (2017) Tissue-specific ubiquitination by IPA1 INTERACTING PROTEIN1 modulates IPA1 protein levels to regulate Plant Architecture in Rice. Plant Cell 29:697–707. https://doi.org/10.1105/tpc.16.00879
Kang K, Shim Y, Gi E et al (2019) Mutation of ONAC096 enhances Grain Yield by increasing panicle number and delaying Leaf Senescence during Grain Filling in Rice. Int J Mol Sci 20:5241. https://doi.org/10.3390/ijms20205241
Kim I-S, Kim Y-S, Yoon H-S (2013) Expression of salt-induced 2-Cys peroxiredoxin from Oryza sativa increases stress tolerance and fermentation capacity in genetically engineered yeast Saccharomyces cerevisiae. Appl Microbiol Biotechnol 97:3519–3533. https://doi.org/10.1007/s00253-012-4410-8
Redillas MCFR, Jeong JS, Kim YS et al (2012) The overexpression of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnol J 10:792–805. https://doi.org/10.1111/j.1467-7652.2012.00697.x
Kusumi K, Hashimura A, Yamamoto Y et al (2017) Contribution of the S-type Anion Channel SLAC1 to Stomatal Control and its dependence on Developmental Stage in Rice. Plant Cell Physiol 58:2085–2094. https://doi.org/10.1093/pcp/pcx142
Wu T, Ali A, Wang J et al (2021) A homologous gene of OsREL2/ASP1, ASP-LSL regulates pleiotropic phenotype including long sterile lemma in rice. BMC Plant Biol 21:390. https://doi.org/10.1186/s12870-021-03163-7
Bae K-D, Um T-Y, Yang W-T et al (2021) Characterization of dwarf and narrow leaf (dnl-4) mutant in rice. Plant Signal Behav 16:1849490. https://doi.org/10.1080/15592324.2020.1849490
Funding
This research was supported by grants from the Department of Science and Technology-Science and Engineering Research Board (DST-SERB), Government of India (Grant No. ECR/2016/001934).
Author information
Authors and Affiliations
Contributions
AM conceived and supervised the research work; MP prepared genetic materials; AM acquired research grant for the research; KTS, MP performed data analyses; MP wrote the manuscript; All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interest to disclose.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Peringottillam, M., Sundaram, K.T. & Manickavelu, A. Genetic potential of grain-related traits in rice landraces: phenomics and multi-locus association analyses. Mol Biol Rep 50, 9323–9334 (2023). https://doi.org/10.1007/s11033-023-08807-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08807-5