Abstract
North Eastern part of India such as Assam is inundated by flood every year where the farmers are forced to grow the traditional tall deep-water rice. Genetic improvement of this type of rice is slow because of insufficient knowledge about their genetic architecture and population structure. In the present investigation, the genetic diversity architecture of 94 deep-water rice genotypes of Assam and association mapping strategy was, for the first time, applied to determine the significant SNPs and genes for deep-water rice. These genotypes are known for their unique elongation ability under deep-water condition. The anaerobic germination (AG) related trait-associated genes identified here can provide affluent resources for rice breeding especially in flood-prone areas. We investigated the genome-wide association studies (GWAS) using 50 K rice genic SNP chip across 94 deep-water rice genotypes collected from different flood-prone districts/villages of Assam. Population structure and diversity analysis revealed that these genotypes were stratified into four sub-populations. Using GWAS approach, 20 significant genes were identified and found to be associated with AG-related traits. Of them, two most relevant genes (OsXDH1and SSXT) have been identified which explain phenotypic variability (R2 > 20%) in the population. These genes were located in Chr 3 (LOC_Os03g31550) which encodes for enzyme xanthine dehydrogenase 1(OsXDH1) and in Chr 12 (LOC_Os12g31350) which encodes for SSXT family protein. Both of these genes were found to be associated with anaerobic response index (increase in the coleoptile length under water in anaerobic condition with respect to control), respectively. Interestingly, OsXDH1is involved in purine catabolism pathway and acts as a scavenger of reactive oxygen species in plants, whereas SSXT is GRF1-interacting factor 3. These two candidate genes associated with AG of deep-water rice have been found to be reported for the first time. Thus, this study provides a greater resource for breeders not only for improvement of deep-water rice, but also for AG tolerant variety useful for direct-seeded rice in flood-affected areas.










Similar content being viewed by others
Availability of data and material
All the data are available in public.
References
Angaji SA, Septiningsih EM, Mackill DJ, Ismail AM (2010) QTLs associated with tolerance of flooding during germination in rice (Oryza sativa L.). Euphytica 172(2):159–168
Baltazar MD, Ignacio JCI, Thomson MJ, Ismail AM, Mendioro MS, Septiningsih EM (2014) QTL mapping for tolerance of anaerobic germination from IR64 and the aus landrace Nanhi using SNP genotyping. Euphytica 197(2):251–260
Baltazar MD, Ignacio JCI, Thomson MJ, Ismail AM, Mendioro MS, Septiningsih EM (2019) QTL mapping for tolerance to anaerobic germination in rice from IR64 and the Aus landrace Kharsu 80A. Breed Sci 69(2):227–233
Bhuyan N, Borah BK, Sarma RN (2007) Genetic diversity analysis in traditional lowland rice (Oryza sativa L.) of Assam using RAPD and ISSR markers. Curr Sci 93:967–997
Biselli C, Volante A, Desiderio F, Tondelli A, Gianinetti A, Finocchiaro F, Taddei F, Gazza L, Sgrulletta D, Cattivelli L, Vale G (2019) GWAS for stach-related parameters in japonica rice (Oryza sativa L.). Plants 8(8):292
Biswas JK, Yamauchi M (1997) Mechanism of seedling establishment of direct-seeded rice (Oryza sativa L.) under lowland conditions. Bot Bull Acad Sin 38:29–32
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: Software for association mapping of complex traits in diverse samples. Bioinform 23:2633–2635
Catling D (1992) Rice in deep-water. International Rice Research Institute, pp 419–426
Chang CL, Serapion JC, Hung HH, Lin YC, Tsai YC, Jane WN, Chang MC, Lai MH, Yue-ie CH (2019) Studies of a rice sterile mutant sstl from the TRIM collection. Bot Stud 60(1):1–6
Choudhury B, Khan ML, Dayanandan S (2013) Genetic structure and diversity of indigenous rice (Oryza sativa) varieties in the Eastern Himalayan region of Northeast India. Springer Plus 2(1):228
Das B, Sengupta S, Parida SK, Roy B, Ghosh M, Prasad M, Ghose TK (2013) Genetic diversity and population structure of rice landraces from Eastern and North Eastern States of India. BMC Genet 14:71–85
Doyle JJ (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Earl DA, Vonholdt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Feng Y, Lu Q, Zhai R, Zhang M, Xu Q, Yang Y, Wang S, Yuan X, Yu H, Wang Y (2016) Genome wide association mapping for grain shape traits in indica rice. Planta 244(4):819–830
Fuller DQ, Sato YI, Castillo C, Qin L, Weisskopf AR, Kingwel-lBanham EJ (2010) Consilience of genetics and archaeobotany in the entangled history of rice. Archaeol Anthropol Sci 2:115–131
Ganie SA, Pani DR, Mondal TK (2017) Genome-wide analysis of DUF221 domain-containing gene family in Oryza species and identification of its salinity stress-responsive members in rice. PLoS ONE 12(8):e0182469
Ghosal S, Casal C, Quilloy FA, Septiningsih EM, Mendioro MS, Dixit S (2019) Deciphering genetics underlying stable anaerobic germination in rice: phenotyping, QTL identification, and interaction analysis. Rice 12(1):50
Goswami DC (1985) Brahmaputra River, Assam, India: Physiography, basin denudation, and channel aggradation. Water Resour Res 21:959–978
Hofmann NR (2016) Opposing functions for plant xanthine dehydrogenase in response to powdery mildew infection: production and scavenging of reactive oxygen species. Plant Cell 28:1001
Hsu SK, Tung CW (2015) Genetic mapping of anaerobic germination-associated QTLs controlling coleoptile elongation in rice. Rice 8(1):38
Huang X, Kurata N, Wei X, Wang ZX, Wang A, Zhao Q, Zhao Y, Liu K, Lu H, Li W, Guo Y, Lu Y, Zhou C, Fan D, Weng Q, Zhu C, Huang T, Zhang L, Wang Y, Feng L, Furuumi H, Kubo T, Miyabayashi T, Yuan X, Xu Q, Dong G, Zhan Q, Li C, Fujiyama A, Toyoda A, Lu T, Feng Q, Qian Q, Li J, Han B (2012) A map of rice genome variation reveals the origin of cultivated rice. Nature 490:497–501
Ismail AM, Ella ES, Vergara GV, Mackill DJ (2009) Mechanisms associated with tolerance to floodingduring germination and early seedling growth in rice (Oryza sativa). Ann Bot 103(2):197–209
Jackson B, Phool C (2003) Physiological and molecular basis of susceptibility and tolerance of rice plants to complete submergence. Ann Bot 91(2):227–241
Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E (2008) Efficient control of population structure in model organism association mapping. Genetics 178:1709–1723
Kawahara Y, De la BM, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu J, Zhou S, Childs KL, Davidson RM, Lin H, Quesada-Ocampo L, Vaillancourt B, Sakai H, Lee SS, Kim J, Numa H, Itoh T, Buell CR, Matsumoto T (2013) Improvement of the Oryzasativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6:4
Kim MS, Reinke FR (2019) A novel resistance gene for bacterial blight in rice, Xa43(t) identified by GWAS, confirmed by QTL mapping using a bi-parental population. PLoS ONE 14:2
Konchan S, Kono Y (1996) Spread of direct seeded lowland rice in Northeast Thailand. J Southeast Asian Stud 33(4):523–546
LiLin Y (2018) Package “CMplot.” 7. https://github.com/YinLiLin/R-CMplot. Accessed 17 Sept 2018
Lim MN, Lee SE, Yim HK, Kim JH, Yoon IS, Hwang YS (2013) Differential anoxic expression of sugar-regulated genes reveals diverse interactions between sugar and anaerobic signaling systems in rice. Mol Cells 36:169–176
Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, Gore MA, Buckler E, Zhang Z (2012) GAPIT: genome association and prediction integrated tool. Bioinformatics 28(18):2397–2409
Liu XL (2015) Development of an Iterative Usage of Fixed Effect and Random Effect Models for Powerful and Efficient Genome-Wide Association Studies. Master’s thesis, Huazhong agricultural University, Wuhan
Liu X, Huang M, Fan B, Buckler ES, Zhang Z (2016) Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet 12:e1005767
Magneschi L, Kudahettige RL, Alpi A, Perata P (2009) Expansin gene expression and anoxic coleoptile elongation in rice cultivars. Plant Physiol 166:1576–1580
Mazumder A, Rohilla M, Bisht DS, Krishamurthy SL, Sharma TR, Mondal TK (2020) Identification and mapping of quantitative trait loci (QTL) and epistatic QTL for salinity tolerance at seedling stage in traditional aromatic short grain rice landrace Kolajoha (Oryza sativa L.) of Assam, India. Euphytica 216:75
Miro B, Ismail AM (2013) Tolerance of anaerobic conditions caused by flooding during germination and early growth in rice (Oryza sativa L.). Front Plant Sci 33:523–546
Mustroph A, Lee SC, Oosumi T, Zanetti ME, Yang H, Ma K, Yaghoubi-Masihi A, Fukao T, Bailey-Serres J (2010) Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol 152(3):1484–1500
Nghi KN, Tondelli A, Valè G, Tagliani A, Marè C, Perata P, Pucciariello C (2019) Dissection of coleoptiles elongation in japonica rice under submergence through integrated genome-wide association mapping and transcriptional analyses. Plant Cell Environ 42(6):1832–1846
Norton GJ, Travis AJ, Douglas A, Fairley S, Alves EP, Ruang-Areerate P, Naredo MEB, McNally KL, Hossain M, Islam MR, Price AH (2018) Genome wide association mapping of grain and straw biomass traits in the Rice Bengal and Assam Aus Panel (BAAP) grown under alternate wetting and drying and permanently flooded irrigation. Front Plant Sci 3(9):1223
Oladosu Y, Rafii MY, Arolu F, Chukwu SC, Muhammad I, Kareem I, Salisu MA, Arolu IW (2020) Submergence tolerance in rice: review of mechanism. Breed Future Prospects Sustain 12(4):1632
Peakall R, Smouse P (2012) GenAlEx 65: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinform 28:2537–2539
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genet 155:945–959
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, De Bakker PI, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 8(1):559–575
Reig-Valiente JL, Marques L, Talon M, Domingo C (2018) Genome-wide association study of agronomic traits in rice cultivated in temperate regions. BMC Genom 19:706
Rohilla M, Roy P, ChowdhuryD SKK, Saikia P, Sen P, Singh NK, Mondal TK (2019) Bao Dhan of Assam: organically grown indigenous rice slated to increase farmer’s income. Curr Sci 116:706–708
Roy CD, Singh N, Singh AK, Kumar S, Srinivasan K, Tyagi RK, Ahmad A, Singh NK, Singh R (2014) Analysis of genetic diversity and population structure of rice germplasm from North-Eastern region of India and development of a core germplasm set. PLoS ONE 9:e113094
Sarma RN, Bahar B (2005) Genetic variation of bora rice (glutinous rice) of Assam as revealed by RAPDs. Plant Genet Resour Newsl 144:34–38
Schmittgen TD, Livak KJ (2008) Real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108
Segura V, Vilhjalmsson BJ, Platt A, Korte A, Seren U, Long Q, Nordborg M (2012) An efficient multi-locus mixed model approach for genome-wide association studies in structured populations. Nat Genet 44(7):825–830
Septiningsih EM, Ignacio JC, Sendon PM, Sanchez DL, Ismail AM, Mackill DJ (2013) QTL mapping and confirmation for tolerance of anaerobic conditions during germination derived from the rice landrace Ma-Zhan Red. Theor Appl Genet 126:1357–1366
Singh N, Jayaswal PK, Panda K, Mandal P, Kumar V, Singh B, Mishra S, Singh Y, Singh R, Rai V, Gupta A, Raj Sharma T, Singh NK (2015) Single-copy gene based 50 K SNP chip for genetic studies and molecular breeding in rice. Sci Rep 5:11600
Singh B, Singh N, Mishra S, Tripathi K, Singh BP, Rai V, Singh AK, Singh NK (2018) Morphological and molecular data reveal three distinct populations of Indian wild rice Oryza rufipogon Griff. Species Complex Front Plant Sci 9:123
Tanaka T, Antonio BA, Kikuchi S, Matsumoto T, Nagamura Y, Numa H (2008) The rice annotation project database (RAP-DB). Nucleic Acids Res 36:D1028–D1033
Vairavan S, Siddiq EA, Arunachalam V, Swaminathan MS (1973) A study on the nature of genetic divergence in rice from Assam and North East Himalayas. Theor Appl Genet 43(5):213–221
Volante A, Desiderio F, Tondelli A, Perrini R, Orasen G, Biselli C, Riccardi P, Vattari A, Cavalluzzo D, Urso S, Ben Hassen M (2017) Genome-wide analysis of japonica rice performance under limited water and permanent flooding conditions. Front Plant Sci 8:1862
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93(1):77–78
Watanabe S, Sato M, Sawada Y, Tanaka M, Matsui A, Kanno Y, Hirai MY, Seki M, Sakamoto A, Seo M (2018) Arabidopsis molybdenum cofactor sulfurase ABA3 contributes to anthocyanin accumulation and oxidative stress tolerance in ABA dependent and independent ways. Sci Rep 8:16592
Yamauchi M, Chuong PV (1995) Rice seedling establishment as affected by cultivar, seed coating with calcium peroxide, sowing depth, and water level. Field Crops Res 41(2):123–134
Yang J, Sun K, Li D, Luo L, Liu Y, Huang M, Yang G, Liu H, Wang H, Chen Z, Guo T (2019) Identification of stable QTLs and candidate genes involved in anaerobic germination tolerance in rice via high-density genetic mapping and RNA-Seq. BMC Genom 20:355
Yim HK, Lim MN, Lee SE, Lim J, Lee Y, Hwang YS (2012) Hexokinase-mediated sugar signaling controls expression of the calcineurin B-Like interacting protein kinase 15 gene and is perturbed by oxidative phosphorylation inhibition. J Plant Physiol 169:1551–1558
Yu J, Pressoir G, Briggs WH, Vroh Bi I, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, Kresovich S, Buckler ES (2005) Aunifiedmixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38:203–208
Yu J, Holland JB, McMullen MD, Buckler ES (2008) Genetic design and statistical power of nested association mapping in maize. Genetics 178(1):539–551
Zhang Z, Ersoz E, Lai CQ, Todhunter RJ, Tiwari HK, Gore MA, Bradbury PJ, Yu J, Arnett DK, Ordovas JM, Buckler ES (2010) Mixed linear model approach adapted for genome-wide association studies. Nat Genet 42:355–360
Zhang M, Lu Q, Wu W, Niu X, Wang C, Feng Y, Xu Q, Wang S, Yuan X, Yu H, Wang Y (2017) Association mapping reveals novel genetic loci contributing to flooding tolerance during germination in indica rice. Front Plant Sci 8:678
Acknowledgements
The authors are grateful to Department of Biotechnology, New Delhi for financial assistance through DBT-TWIN mode project. The authors are also grateful to Director, ICAR-National Institute for Plant Biotechnology, New Delhi, for providing the facility and. Mr. Mahammed Saba Rahim to provide valuable suggestions for some part of the statistical analysis
Funding
The work was funded by Department of Biotechnology, Govt of India, New Delhi, through a competitive grant number: BT/PR16907/NER/95/345/2015.
Author information
Authors and Affiliations
Contributions
MR did most of the wet lab experiment, NS did most of the statistical analysis, AM did part of the wet lab experiment and part of statistical analysis, DC did most of the experiment, supplied the seeds of all the genotypes, PS did part of the wet lab experiment, NKS guided the work, and TKM conceived the experiment, wrote majority of the manuscript which was assisted by MR, NS, and AM.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
All the authors have consent for publication.
Additional information
Communicated by S. Hohmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
438_2020_1690_MOESM1_ESM.pptx
Suppl.Fig.S1:Diversity analysis in deep-water rice collections: a) Estimation of rate of heterozygosity using 50K SNP markers, x-axis=Heterozygosity rate, y-axis= number of SNPs, b) Estimation of minor allele frequency (MAF): x-axis=MAF Value, y-axis=number of SNPs (PPTX 56 kb)
438_2020_1690_MOESM2_ESM.pptx
Suppl.Fig.S2: Frequency distribution of SNPs variation in duplicated names of deep-water rice genotypes used in the present study (PPTX 71 kb)
438_2020_1690_MOESM3_ESM.pptx
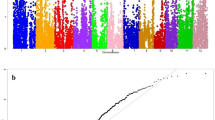
Suppl.Fig.S3: Genome wide Manhattan plots and Q/Q plots of association mapping for AG using GLM: a) Q/Q plots for germination percentage b) Manhattan plots for germination percentage c) Q/Q plots for survival percentage d) Manhattan plots for survival percentage e) Q/Q plots for anaerobic response index f) Manhattan plots for anaerobic response index. (PPTX 1351 kb)
438_2020_1690_MOESM4_ESM.pptx
Suppl.Fig.S4: Genome wide Manhattan plots and Q/Q plots of association mapping for AG using MLMM: a) Q/Q plots for germination percentage b) Manhattan plots for germination percentage c) Q/Q plots for survival percentage d) Manhattan plots for survival percentage e) Q/Q plots for anaerobic response index f) Manhattan plots for anaerobic response index. (PPTX 1336 kb)
438_2020_1690_MOESM5_ESM.pptx
Suppl.Fig.S5: Genome wide Manhattan plots and Q/Q plots of association mapping for AG using FarmCPU: a) Q/Q plots for germination percentage b) Manhattan plots for germination percentage c) Q/Q plots for survival percentage d) Manhattan plots for survival percentage e) Q/Q plots for anaerobic response index f) Manhattan plots for anaerobic response index. (PPTX 1092 kb)
Rights and permissions
About this article
Cite this article
Rohilla, M., Singh, N., Mazumder, A. et al. Genome-wide association studies using 50 K rice genic SNP chip unveil genetic architecture for anaerobic germination of deep-water rice population of Assam, India. Mol Genet Genomics 295, 1211–1226 (2020). https://doi.org/10.1007/s00438-020-01690-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-020-01690-w