Abstract
Background
ToxA, a necrotrophic effector protein, is present in the genome of fungal species like Parastagnospora nodorum, Pyrenophora tritici-repentis and Bipolaris sorokiniana. Tsn1 is the sensitivity gene in the host whose presence indicates more susceptibility to ToxA carrying pathogen, and ToxA-Tsn1 interaction follows an inverse gene-for-gene relationship.
Methods and results
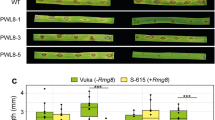
The present study involved cloning and expressing the ToxA1 haplotype from B. sorokiniana. It was found that the amplicon exhibited an expected product size of 471 bp. Sequence analysis of the ToxA1 nucleotide sequence revealed the highest identity, 99.79%, with P. tritici-repentis. The protein expression analysis showed peak expression at 16.5 kDa. Phylogenetic analysis of the ToxA1 sequence from all the Bipolaris isolates formed an independent clade along with P. tritici-repentis and diverged from P. nodorum. ToxA-Tsn1 interaction was studied in 18 wheat genotypes (11 Tsn1 and 7 tsn1) at both seedling and adult stages, validating the inverse gene-for-gene relationship, as the toxin activity was highest in the K68 genotype (Tsn1) and lowest in WAMI280 (tsn1).
Conclusion
The study indicates that the haplotype ToxA1 is prevailing in the Indian population of B. sorokiniana. It would be desirable for wheat breeders to select genotypes with tsn1 locus for making wheat resistant to spot blotch.






Similar content being viewed by others
Data availability
Not applicable.
References
Ciuffetti LM, Tuori RP, Gaventa JM (1997) A single gene encodes a selective toxin causal to the development of tan spot of wheat. Plant Cell 9:135–144. https://doi.org/10.1105/tpc.9.2.135
Wolpert TJ, Dunkle LD, Ciuffetti LM (2002) Host-selective toxins and avirulence determinants: what’s in a name? Annu Rev Phytopathol 40:251–285. https://doi.org/10.1146/annurev.phyto.40.011402.114210
Sharma P, Duveiller E, Sharma RC (2006) Effect of mineral nutrients on spot blotch severity in wheat and associated increases in grain yield. Field Crops Res 95:426–430. https://doi.org/10.1016/j.fcr.2005.04.015
Pandey AK, Mishra VK, Chand R, Navathe S, Budhlakoti N, Srinivasa J, Sharma S, Joshi AK (2021) Crosses with spelt improve tolerance of South Asian spring wheat to spot blotch, terminal heat stress, and their combination. Sci Rep 11:1–12. https://doi.org/10.1038/s41598-021-85238-x
Ballance GM, Lamari L, Bernier CC (1989) Purification and characterization of a host-selective necrosis toxin from Pyrenophora tritici-repentis. Physiol Mol Plant Pathol 35:203–213. https://doi.org/10.1016/0885-5765(89)90051-9
Friesen TL, Stukenbrock EH, Liu Z, Meinhardt S, Ling H, Faris JD, Oliver, R. P. (2006) Emergence of a new disease as a result of interspecific virulence gene transfer. Nat. Genet. 38:953–956. https://doi.org/10.1038/ng1839
McDonald MC, Solomon PS (2018) Just the surface: advances in the discovery and characterization of necrotrophic wheat effectors. Curr Opin Microbiol 46:14–18
Solomon P, Lowe R, Tan KC, Waters O, Oliver R (2006) Stagonospora nodorum; cause of Stagonospora nodorum blotch of wheat. Mol Plant Pathol 7:147–156. https://doi.org/10.1111/j.1364-3703.2006.00326.x
Tuori RP, Wolpert TJ, Ciuffetti LM (1995) Purification and immunological characterization of toxic components from cultures of Pyrenophora tritici-repentis. Mol Plant Microbe Interact 8:41–48. https://doi.org/10.1094/mpmi-8-0041
Tomás A, Bockus WW (1987) Cultivar specific toxicity of culture filtrate of Pyrenophora tritici-repentis. Phytopathology 77:1337–1366. https://doi.org/10.1094/phyto-77-1337
McDonald MC, Ahren D, Simpfendorfer S, Milgate A, Solomon PS (2018) The discovery of the virulence gene ToxA in the wheat and barley pathogen Bipolaris sorokiniana. Mol Plant Pathol 19:432–439. https://doi.org/10.1111/mpp.12535
Ballance GM, Lamari L, Kowatsch R, Bernier CC (1996) Cloning, expression and occurrence of the gene encoding the Ptr necrosis toxin from Pyrenophora tritici-repentis. Mol Plant Pathol. https://doi.org/10.1007/978-94-011-5218-1_21
Tuori RP, Wolpert TJ, Ciuffetti LM (2000) Heterologous expression of functional PtrToxA. Mol Plant Microbe Interact 13:456–464. https://doi.org/10.1094/mpmi.2000.13.4.456
Faris JD, Zhang Z, Lu H, Lu S, Reddy L, Cloutier S, Friesen, T. L. (2010) A unique wheat disease resistance-like gene governs effector-triggered susceptibility to necrotrophic pathogens. Proc Natl Acad Sci USA 107:13544–13549. https://doi.org/10.1073/pnas.1004090107
Lamari L, Bernier CC (1989) Evaluation of wheat lines and cultivars to tan spot [Pyrenophora tritici-repentis] based on lesion type. Can J Plant Pathol 11:49–56. https://doi.org/10.1080/07060668909501146
Tan KC, Ferguson-Hunt M, Rybak K, Waters OD, Stanley WA, Bond CS, Stukenbrock EH, Friesen TL, Faris JD, McDonald BA, Oliver RP (2012) Quantitative variation in effector activity of ToxA isoforms from Stagonospora nodorum and Pyrenophora tritici-repentis. Mol Plant Microbe Interact 25:515–522. https://doi.org/10.1094/mpmi-10-11-0273
Sambrook J, Russell DW, Sambrook J (2006) The condensed protocols: from molecular cloning: a laboratory manual, (No. Sirsi) i9780879697723). Cold Spring Harb Protoc NY. https://doi.org/10.1101/pdb.prot4018
Hall TA (1999) A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. https://doi.org/10.1093/nar/25.17.3389
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. https://doi.org/10.1093/bioinformatics/btm404
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. https://doi.org/10.1093/bioinformatics/8.3.275
Felsenstein, (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODELworkspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:95–201. https://doi.org/10.1093/bioinformatics/bti770
Benkert P, Tosatto SCE, Schwede T (2009) Global and local model quality estimation at CASP8 using the scoring functions QMEAN and QMEANclust. Proteins Struct Funct Bioinforma 77:173–180. https://doi.org/10.1002/prot.22532
Studer G, Rempfer C, Waterhouse AM, Gumienny R, Haas J, Schwede T (2020) QMEAND is Co—distance constraints applied on model quality estimation. Bioinformatics 36:1765–1771. https://doi.org/10.1093/bioinformatics/btz828
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291. https://doi.org/10.1107/s0021889892009944
Tian W, Chen C, Lei X, Zhao J, Liang J (2018) CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res 46:W363–W367. https://doi.org/10.1093/nar/gky473
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera. A visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. https://doi.org/10.1002/jcc.20084
Zadoks JC, Chang TT, Konzak CF (1974) A decimal code for the growth stages of cereals. Weed Res 14:415–421. https://doi.org/10.1111/j.1365-3180.1974.tb01084.x
Abramoff MD, Magalhaes PJ, Ram SJ (2004) Image processing with ImageJ. Biophotonics Int 11:36–42
Tate RF (1954) Correlation between a discrete and a continuous variable. Point-biserial correlation. Ann Math Stat 25:603–607
R Core Team (2022) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
Navathe S, Yadav PS, Chand R, Mishra VK, Vasistha NK, Meher PK, Joshi AK, Gupta PK (2020) ToxA–Tsn1 interaction for spot blotch susceptibility in Indian wheat: an example of inverse gene-for-gene relationship. Plant Dis 104:71–81. https://doi.org/10.1094/pdis-05-19-1066-re
McDonald MC, Razavi M, Friesen TL, Brunner PC, McDonald BA (2012) Phylogenetic and population genetic analyses of Phaeosphaeria nodorum and its close relatives indicate cryptic species and an origin in the Fertile Crescent. Fungal Genet Biol 49:882–895. https://doi.org/10.1016/j.fgb.2012.08.001
Manning VA, Hamilton SM, Karplus PA, Ciuffetti LM (2008) The Arg-Gly-Asp–containing, solvent-exposed loop of PtrToxA is required for internalization. Mol Plant Microbe Interact 21:315–325. https://doi.org/10.1094/mpmi-21-3-0315
Hafez M, Despins T, Nakajima K, Aboukhaddour R (2022) Identification of a novel ToxA haplotype of Pyrenophora tritici-repentis from Japan. Phytopathology 112:1597–1602. https://doi.org/10.1094/phyto-01-22-0001-sc
Aboukhaddour R, Abdel-Fattah MH, McDonald M, Moffat C, Navathe S, Friesen TL, Strelkov SE, Oliver RP, Tan K-C, Liu Z, Moolhuijzen P, Phan H, See PT, Solomon P (2023) A revised nomenclature for ToxA haplotypes across multiple fungal species. Phytopathology. https://doi.org/10.1094/phyto-01-23-0017-sc
Sarma GN, Manning VA, Ciuffetti LM, Karplus PA (2005) Structure of PtrToxA: an RGD-containing host-selective toxin from Pyrenophora tritici-repentis. Plant Cell 17:3190–3202. https://doi.org/10.1105/tpc.105.034918
Ciuffetti LM, Manning VA, Pandelova I, Betts MF, Martinez JP (2010) Host-selective toxins, PtrToxA and PtrToxB, as necrotrophic effectors in the Pyrenophora tritici-repentis–wheat interaction. New Phytol 187:911–919. https://doi.org/10.1111/j.1469-8137.2010.03362.x
McDonald MC, Solomon PS (2018) Just the surface: advances in the discovery and characterization of necrotrophic wheat effectors. Curr Opin Microbiol 46:14–18. https://doi.org/10.1016/j.mib.2018.01.019
Manning VA, Hardison LK, Ciuffetti LM (2007) PtrToxA interacts with a chloroplast-localized protein. Mol Plant Microbe Interact 20:168–177. https://doi.org/10.1094/mpmi-20-2-0168
Aggarwal R, Agarwal S, Sharma S, Gurjar MS, Bashyal BM, Rao AR, Saharan MS (2021) Whole genome sequencing and expression analysis of ToxA in Bipolaris sorokiniana provides discernment of pathogenicity causing spot blotch of wheat. Preprint (Version 1) available at Research Square. https://doi.org/10.21203/rs.3.rs-677095/v1
Virdi S, Liu Z, Overlander M, Zhang Z, Xu SS, Friesen T, Faris JD (2016) New insights into the roles of host gene-necrotrophic effector interactions in governing susceptibility of durum wheat to tan spot and Septoria nodorum blotch. Genes Genomes Genet 6:4139–4150. https://doi.org/10.1534/g3.116.036525
Faris JD, Liu Z, Xu SS (2013) Genetics of tan spot resistance in wheat. Theor Appl Genet 126:2197–2217. https://doi.org/10.1007/s00122-013-2157-y
Acknowledgements
Authors acknowledge Banaras Hindu University and Bihar Agricultural University for the infrastructure facility
Funding
SN acknowledges SERB, New Delhi, for CRG/2021/007211.
Author information
Authors and Affiliations
Contributions
Conceptualization, RC, VKM, PKS, SN, and SK; methodology, RKC and DT; validation, DT and SS; formal analysis, DT and SS; investigation, RKC; resources, RC, DT; data curation, DT, SS; writing—original draft preparation, RKC, SN; writing—review and editing, RC, PKS; supervision, RC, VKM, SS; project administration, RC; funding acquisition, RC, PKS. All authors have read and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Ethical approval
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chaubey, R.K., Thakur, D., Navathe, S. et al. Heterologous expression and characterization of ToxA1 haplotype from India and its interaction with Tsn1 for spot blotch susceptibility in spring wheat. Mol Biol Rep 50, 8213–8224 (2023). https://doi.org/10.1007/s11033-023-08717-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08717-6