Abstract
Background
Sugars produced by photosynthesis provide energy for biological activities and the skeletons for macromolecules; they also perform multiple physiological functions in plants. Sugar transport across plasma membranes mediated by the Sugar Will Eventually be Exported Transporter (SWEET) genes substantially affects these processes. However, the evolutionary dynamics and function of the SWEET genes are largely unknown in radish, an important Brassicaceae species.
Methods and results
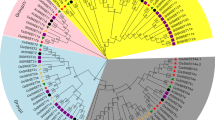
Genome-wide identification and analysis of the RsSWEET genes from the recently updated radish reference genome was conducted using bioinformatics methods. The tissue-specific expression was analyzed using public RNA-seq data, and the expression levels in the bud, stamens, pistils, pericarps and seeds at 15 and 30 days after flowering (DAF) were determined by RT‒qPCR. Thirty-seven RsSWEET genes were identified and named according to their Arabidopsis homologous. They are unevenly distributed across the nine radish chromosomes and were further divided into four clades by phylogenetic analysis. There are 5–7 transmembrane domains and at least one MtN3_slv domain in the RsSWEETs. RNA-seq and RT‒qPCR revealed that the RsSWEETs exhibit higher expression levels in the reproductive organs, indicating that these genes might play vital roles in reproductive organ development. RsSWEET15.1 was found to be especially expressed in siliques according to the RNA-seq data, and the RT‒qPCR results further confirmed that it was most highly expressed levels in the seeds at 30 DAF, followed by the pericarp at 15 DAF, indicating that it is involved in seed growth and development.
Conclusions
This study suggests that the RsSWEET genes play vital roles in reproductive organ development and provides a theoretical basis for the future functional analysis of RsSWEETs in radish.





Similar content being viewed by others
Data availability
The R. sativus ‘Xin-li-mei’ whole-genome sequence was downloaded from the CNCB database with accession number GWHANWD00000000 (https://ngdc.cncb.ac.cn/gwh/Assembly/9797/show). The RNA-seq reads in different tissues (root, leaf, bolting tissue, flower, silique and callus) of R. sativus are available at the NCBI Sequence Read Archive (SRA, http://www.ncbi.nlm.nih.gov/Traces/sra) (PRJNA413464).
References
Walmsley AR, Barrett MP, Bringaud F, Gould GW (1998) Sugar transporters from bacteria, parasites and mammals: structure–activity relationships. Trends Biochem Sci 23(12):476–481
Neuhaus HE (2007) Transport of primary metabolites across the plant vacuolar membrane. FEBS Lett 581(12):2223–2226
Rolland F, Baena-Gonzalez E, Sheen J (2006) Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol 57(1):675–709
Griffiths CA, Paul MJ, Foyer CH (1857) Metabolite transport and associated sugar signalling systems underpinning source/sink interactions. BBA-Bioenergetics 10:1715–1725
Eom JS, Chen LQ, Sosso D, Julius BT, Lin IW, Qu XQ, Braun DM, Frommer WB (2015) SWEETs, transporters for intracellular and intercellular sugar translocation. Curr Opin Plant Biol 25:53–62
Buttner M (2010) The Arabidopsis sugar transporter (AtSTP) family: an update. Plant Biol 12(Suppl 1):35–41
Chen LQ (2014) SWEET sugar transporters for phloem transport and pathogen nutrition. New Phytol 201(4):1150–1155
Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, Qu XQ, Guo WJ, Kim JG, Underwood W, Chaudhuri B (2010) Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468(7323):527–532
Yuan M, Wang S (2013) Rice MtN3/Saliva/SWEET family genes and their homologs in cellular organisms. Mol Plant 6(3):665–674
Filyushin MA, Anisimova OK, Shchennikova AV, Kochieva EZ (2023) Genome-wide identification, expression, and response to Fusarium infection of the SWEET gene family in garlic (Allium sativum L.). Int J Mol Sci 24(8):7533
Patil G, Valliyodan B, Deshmukh R, Prince S, Nicander B, Zhao M, Sonah H, Song L, Lin L, Chaudhary J et al (2015) Soybean (Glycine max) SWEET gene family: insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis. BMC Genomics 16:520
Li M, Xie H, He M, Su W, Yang Y, Wang J, Ye G, Zhou Y (2020) Genome-wide identification and expression analysis of the StSWEET family genes in potato (Solanum tuberosum L.). Genes Genom 42(2):135–153
Chong J, Piron MC, Meyer S, Merdinoglu D, Bertsch C, Mestre P (2014) The SWEET family of sugar transporters in grapevine: VvSWEET4 is involved in the interaction with Botrytis cinerea. J Exp Bot 65(22):6589–6601
Xuan C, Lan G, Si F, Zeng Z, Wang C, Yadav V, Wei C, Zhang X (2021) Systematic genome-wide study and expression analysis of sweet gene family: sugar transporter family contributes to biotic and abiotic stimuli in watermelon. Int J Mol Sci 22(16):8407
Li H, Li X, Xuan Y, Jiang J, Wei Y, Piao Z (2018) Genome wide identification and expression profiling of SWEET genes family reveals its role during Plasmodiophora brassicae-induced formation of clubroot in Brassica rapa. Front Plant Sci 9:207
Hu LP, Zhang F, Song SH, Tang XW, Xu H, Liu GM, Wang Y, He HJ (2017) Genome-wide identification, characterization, and expression analysis of the SWEET gene family in cucumber. J Integr Agr 16(7):1486–1501
Breia R, Conde A, Badim H, Fortes AM, Geros H, Granell A (2021) Plant SWEETs: from sugar transport to plant-pathogen interaction and more unexpected physiological roles. Plant Physiol 186(2):836–852
Ni J, Li J, Zhu R, Zhang M, Qi K, Zhang S, Wu J (2020) Overexpression of sugar transporter gene PbSWEET4 of pear causes sugar reduce and early senescence in leaves. Gene 743:144582
Seo PJ, Park J-M, Kang SK, Kim S-G, Park C-M (2011) An Arabidopsis senescence-associated protein SAG29 regulates cell viability under high salinity. Planta 233(1):189–200
Liu X, Zhang Y, Yang C, Tian Z, Li J (2016) AtSWEET4, a hexose facilitator, mediates sugar transport to axial sinks and affects plant development. Sci Rep 6(1):1–12
Bezrutczyk M, Hartwig T, Horschman M, Char SN, Yang J, Yang B, Frommer WB, Sosso D (2018) Impaired phloem loading in zmsweet13a, b, c sucrose transporter triple knock-out mutants in Zea mays. New Phytol 218(2):594–603
Yu Y, Streubel J, Balzergue S, Champion A, Boch J, Koebnik R, Feng J, Verdier V, Szurek B (2011) Colonization of rice leaf blades by an african strain of Xanthomonas oryzae pv. oryzae depends on a new TAL effector that induces the rice Nodulin-3 Os11N3 Gene. Mol Plant Microbe In 24(9):1102–1113
Liu Q, Yuan M, Zhou Y, Li X, Xiao J, Wang S (2011) A paralog of the MtN3/saliva family recessively confers race-specific resistance to Xanthomonas oryzae in rice. Plant Cell Environ 34(11):1958–1969
Jeena GS, Kumar S, Shukla RK (2019) Structure, evolution and diverse physiological roles of SWEET sugar transporters in plants. Plant Mol Biol 100(4):351–365
Gao Y, Zhang C, Han X, Wang ZY, Ma L, Yuan DP, Wu JN, Zhu XF, Liu JM, Li DP et al (2018) Inhibition of OsSWEET11 function in mesophyll cells improves resistance of rice to sheath blight disease. Mol Plant Pathol 19(9):2149–2161
Quirino BF, Normanly J, Amasino RM (1999) Diverse range of gene activity during Arabidopsis thaliana leaf senescence includes pathogen-independent induction of defense-related genes. Plant Mol Biol 40(2):267–278
Le Hir R, Spinner L, Klemens PA, Chakraborti D, de Marco F, Vilaine F, Wolff N, Lemoine R, Porcheron B, Géry C (2015) Disruption of the sugar transporters AtSWEET11 and AtSWEET12 affects vascular development and freezing tolerance in Arabidopsis. Mol Plant 8(11):1687–1690
Zhang RNK, Ma H (2020) Identification and expression analysis of the SWEET gene family from Poa pratensis under abiotic stresses. DNA Cell Biol 39(9):1606–1620
Kanno Y, Oikawa T, Chiba Y, Ishimaru Y, Shimizu T, Sano N, Koshiba T, Kamiya Y, Ueda M, Seo M (2016) AtSWEET13 and AtSWEET14 regulate gibberellin-mediated physiological processes. Nat Commun 7(1):13245
Morii M, Sugihara A, Takehara S, Kanno Y, Kawai K, Hobo T, Hattori M, Yoshimura H, Seo M, Ueguchi-Tanaka M (2020) The dual function of OsSWEET3a as a gibberellin and glucose transporter is important for young shoot development in rice. Plant Cell Physiol 61(11):1935–1945
Li Y, Liu H, Yao X, Wang J, Feng S, Sun L, Ma S, Xu K, Chen L-Q, Sui X (2021) Hexose transporter CsSWEET7a in cucumber mediates phloem unloading in companion cells for fruit development. Plant Physiol 186(1):640–654
Guan YF, Huang XY, Zhu J, Gao JF, Zhang HX, Yang ZN (2008) RUPTURED POLLEN GRAIN1, a member of the MtN3/saliva gene family, is crucial for exine pattern formation and cell integrity of microspores in Arabidopsis. Plant Physiol 147(2):852–863
Guo C, Li H, Xia X, Liu X, Yang L (2018) Functional and evolution characterization of SWEET sugar transporters in Ananas comosus. Biochem Bioph Res Co 496(2):407–414
Zhen Q, Fang T, Peng Q, Liao L, Zhao L, Owiti A, Han Y (2018) Developing gene-tagged molecular markers for evaluation of genetic association of apple SWEET genes with fruit sugar accumulation. Hortic Res 5(1):1–12
Chen LQ, Lin IW, Qu XQ, Sosso D, McFarlane HE, Londoño A, Samuels AL, Frommer WB (2015) A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo. Plant Cell 27(3):607–619
Yang J, Luo D, Yang B, Frommer WB, Eom JS (2018) SWEET11 and 15 as key players in seed filling in rice. New Phytol 218(2):604–615
Fei H, Yang Z, Lu Q, Wen X, Zhang Y, Zhang A, Lu C (2021) OsSWEET14 cooperates with OsSWEET11 to contribute to grain filling in rice. Plant Sci 306:110851
Sosso D, Luo D, Li QB, Sasse J, Yang J, Gendrot G, Suzuki M, Koch KE, McCarty DR, Chourey PS et al (2015) Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat Genet 47(12):1489–1493
Wang S, Yokosho K, Guo R, Whelan J, Ruan YL, Ma JF, Shou H (2019) The soybean sugar transporter GmSWEET15 mediates sucrose export from endosperm to early embryo. Plant Physiol 180(4):2133–2141
Xie H, Wang D, Qin Y, Ma A, Fu J, Qin Y, Hu G, Zhao J (2019) Genome-wide identification and expression analysis of SWEET gene family in Litchi chinensis reveal the involvement of LcSWEET2a/3b in early seed development. BMC Plant Biol 19(1):1–13
Song J-M, Guan Z, Hu J, Guo C, Yang Z, Wang S, Liu D, Wang B, Lu S, Zhou R et al (2020) Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat Plants 6(1):34–45
Liu T, Zhang Y, Zhang X, Sun Y, Wang H, Song J, Li X (2019) Transcriptome analyses reveal key genes involved in skin color changes of ‘Xinlimei’ radish taproot. Plant Physiol Biochem 139:528–539
Zhang X, Liu T, Wang J, Wang P, Qiu Y, Zhao W, Pang S, Li X, Wang H, Song J et al (2021) Pan-genome of Raphanus highlights genetic variation and introgression among domesticated, wild, and weedy radishes. Mol Plant 14(12):2032–2055
Finn RD, Clements J, Eddy SR (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39:W29-37
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31(13):3784–3788
Chou KC, Shen HB (2010) Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS one 5(6):e11335
Krogh A, Larsson B, Von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305(3):567–580
Tao Y, Cheung LS, Li S, Eom J-S, Chen LQ, Xu Y, Perry K, Frommer WB, Feng L (2015) Structure of a eukaryotic SWEET transporter in a homotrimeric complex. Nature 527(7577):259–263
Wei Y, Xiao D, Zhang C, Hou X (2019) The expanded SWEET gene family following whole genome triplication in Brassica rapa. Genes 10(9):722
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202-208
Chen C, Chen H, Zhang Y, Thomas HR, Xia R (2020) TBtools: Zan integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13(8):1194–1202
Wang J, Qiu Y, Wang X, Yue Z, Yang X, Chen X, Zhang X, Shen D, Wang H, Song J et al (2017) Insights into the species-specific metabolic engineering of glucosinolates in radish (Raphanus sativus L.) based on comparative genomic analysis. Sci Rep 7(1):16040
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25(4):402–408
Gautam T, Saripalli G, Gahlaut V, Kumar A, Sharma P, Balyan H, Gupta P (2019) Further studies on sugar transporter (SWEET) genes in wheat (Triticum aestivum L.). Mol Biol Rep 46(2):2327–2353
Manck-Götzenberger J, Requena N (2016) Arbuscular mycorrhiza symbiosis induces a major transcriptional reprogramming of the potato SWEET sugar transporter family. Front Plant Sci 7:487
Feng CY, Han JX, Han XX, Jiang J (2015) Genome-wide identification, phylogeny, and expression analysis of the SWEET gene family in tomato. Gene 573(2):261–272
Li J, Qin M, Qiao X, Cheng Y, Li X, Zhang H, Wu J (2017) A new insight into the evolution and functional divergence of SWEET transporters in Chinese white pear (Pyrus bretschneideri). Plant Cell Physiol 58(4):839–850
Chu Z, Yuan M, Yao J, Ge X, Yuan B, Xu C, Li X, Fu B, Li Z, Bennetzen JL (2006) Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Gene Dev 20(10):1250–1255
Funding
This study was funded by the National Natural Science Foundation of China (31801858); Research Foundation Incubation Project of Jinling Institute of Technology (jit-fhxm-202113); and Research Foundation for Talented Scholars of Jinling Institute of Technology (jit-b-202009).
Author information
Authors and Affiliations
Contributions
TL: Methodology, Investigation, Formal analysis, Writing-original draft, review & revision, Software, Funding acquisition. QC: Methodology, Analysis, Validation, Writing-original draft. QB: Investigation. LZ: Resources. YY: Investigation. AZ: Investigation. QW: Daily management. CW: Conceptualization, Investigation, Supervision, Project administration.
Corresponding authors
Ethics declarations
Competing interest
The authors declare that they have no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11033_2023_8701_MOESM2_ESM.tif
Supplementary file2 (TIF 1778 KB)—Multiple sequence alignment of the SWEET proteins in radish and A. thaliana SWEET1. The seven TM domains were assigned based on the AtSWEET1 structure indicated by Tao et al. (2015)
11033_2023_8701_MOESM6_ESM.xlsx
Supplementary file6 (XLSX 12 KB)—The fragments per kilobase of transcript per million mapped reads (FPKM) values of the radish SWEET genes in various organs based on RNA-seq data
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, T., Cui, Q., Ban, Q. et al. Identification and expression analysis of the SWEET genes in radish reveal their potential functions in reproductive organ development. Mol Biol Rep 50, 7535–7546 (2023). https://doi.org/10.1007/s11033-023-08701-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08701-0