Abstract
Objective
To detect the expression level of urinary exosomal lncRNA SNHG16 in patients with bladder cancer and healthy individuals and explore its clinical application value in the diagnosis of bladder cancer.
Methods
Urine samples were collected from 42 patients with bladder cancer and 42 healthy volunteers who visited Lu’an Hospital of Anhui Medical University and the Second Hospital of Tianjin Medical University from January 2020 to December 2022. The expression levels of lncRNA SNHG16 in urinary exosomes of the two groups were detected by RT‒qPCR, and their correlation with clinical pathological parameters of bladder cancer patients was analysed. An Receiver Operating Characteristic(ROC) curve was drawn to analyse the diagnostic value of urinary exosomal lncRNA SNHG16 for bladder cancer and compared with urinary cytology.
Results
The expression of urinary exosomal lncRNA SNHG16 in patients with bladder cancer was significantly higher (P < 0.05), and the expression level had no correlation with the age, sex, pathological T stage, pathological grade, or tumour size of bladder cancer patients (P > 0.05). The Area Under Curve(AUC) of urinary exosomal lncRNA SNHG16 in diagnosing bladder cancer was 0.791, which was superior to that of urinary cytology (AUC = 0.597).
Conclusion
Urinary exosomal lncRNA SNHG16 with high expression can serve as a potential diagnostic biological marker for bladder cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Bladder cancer is a highly prevalent malignant tumour worldwide, accounting for 90–95% of urothelial carcinomas. The age-standardized incidence rate of bladder cancer is 9.0 per 100,000 in men and 2.2 per 100,000 in women worldwide [1]. Despite the continuous development of treatment methods, the five-year survival rate for bladder cancer remains less than 20% [2], especially for metastatic bladder cancer [1, 3, 4]. Early diagnosis is the key to timely and effective treatment, improving the prognosis of bladder cancer and reducing the recurrence rate and mortality. Currently, cystoscopy is the gold standard for diagnosing bladder cancer, but due to its complexity and invasiveness, it is difficult to promote it widely in population screening, resulting in a considerable number of patients being diagnosed with muscle-invasive bladder cancer at the first diagnosis, missing the optimal timing for treatment and leading to a poor prognosis. Urine cytology, as one of the most common methods for auxiliary diagnosis of bladder cancer, has a sensitivity of 25-95% for detecting urothelial carcinoma [5,6,7,8,9,10]. However, it is relatively ineffective in identifying low-grade bladder tumours. Imaging methods, such as CT and ultrasound, are easy to use to judge large-volume occupying lesions, but at this point, the patient is already in the middle and late stages, missing the optimal timing for treatment. CT and ultrasound are prone to miss small lesions, which is not conducive to the early detection and treatment of tumours [11, 12]. Therefore, it is urgent to find reliable and noninvasive biomarkers to assist in the early diagnosis of bladder cancer.
LncRNAs are a family of noncoding RNAs defined as transcripts larger than 200 nucleotides. Increasing evidence suggests that the expression of lncRNAs changes in response to tumour development, which can be used for the diagnosis, progression, and prognosis of cancer. However, naked lncRNAs are susceptible to degradation by ribonucleases in bodily fluids and require protection from extracellular vesicles. Exosomes are cell-derived microvesicles with a diameter of approximately 30–150 nm that are present in almost all body fluids. Exosomal vesicles act as “carriers” and “protective shields” that wrap around lncRNAs, preventing their degradation by ribonucleases in bodily fluid circulation [13, 14]. However, the functional contents of these vesicles are not randomly loaded but rather produced differentially according to various pathological conditions [15]. Numerous studies have shown that the expression of lncRNAs in exosomes from the blood or urine of different cancer patients is correspondingly upregulated or downregulated and may be related to tumour staging and prognosis [16,17,18,19,20], making them potential noninvasive biomarkers. SNHG16, as a protumor lncRNA, is upregulated in various human cancers, such as lung cancer, breast cancer, colorectal cancer, and cervical cancer [21,22,23,24,25,26,27]. In this study, we explored the predictive value of urinary exosome-derived lncRNA SNHG16 in the diagnosis of bladder cancer.
Data and methods
Collection of urine specimens
Urinary specimens were collected from 42 patients with bladder cancer and 42 healthy volunteers who underwent physical examinations at Lu’an Hospital of Anhui Medical University and the Second Hospital of Tianjin Medical University between January 2020 and December 2022, defined as the bladder cancer group and control group, respectively. Informed consent was obtained from all participants, and the study was approved by the ethics committee of the Second Hospital of Tianjin Medical University. Inclusion criteria: [1] all subjects were clinically diagnosed with bladder cancer by a physician and confirmed by surgical pathology or cystoscopy biopsy; [2] had no history of radiotherapy, chemotherapy or other anticancer treatments. The exclusion criteria were as follows: [1] patients with other organ dysfunctions, such as heart, lung, liver, or kidney dysfunction; [2] patients with heart disease, diabetes, hypertension, mental disorders, organ failure, immune diseases, and traumatic diseases; and [3] patients with other systemic tumours. Tumour grading and TNM staging were performed according to the WHO staging criteria. After collecting 100 ml of urine sample for each case, preprocessing was performed within two hours, followed by centrifugation at 3000×g for 20 min to obtain the supernatant, which was stored at -80 °C for exosome extraction. The centrifugal precipitate was sent to the pathology department for urine cytology testing.
Cell culture
Human Bladder cancer cell T24 (ATCC) and Benign urothelial cell SV-HUC-1 (ATCC) were cultured, subcultured and cryo-preserved in Tianjin Institute of Urology. All the cells were cultured in a saturated humidity incubator at 37℃ and 5% CO2.
Enrichment of urinary extracellular vesicles
The preprocessed urine samples were subjected to differential centrifugation using an ultrahigh-speed centrifuge (Beckman Coulter, USA) at 4 °C and 17,000 × g for 30 min. The supernatant was collected and subjected to a second centrifugation at 4 °C and 200,000 × g for 70 min. The supernatant was discarded, and 100 µl of PBS was added to resuspend the extracellular vesicles. The resuspended solution was aliquoted and stored at -80 °C for further analysis.
Extraction and reverse transcription of extracellular vesicle RNA
Total RNA was extracted from extracellular vesicles using the Exosome RNA Isolation Kit (Rengen Biosciences Co., Ltd, China). The concentration and integrity of total RNA were evaluated using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Purified RNA was reverse transcribed into cDNA according to the manufacturer’s instructions of the US EVERBRIGHT INC kit. The 20 µl reaction mixture contained 500 ng of template RNA, 4 µl of 5×UEIris II RT MasterMix, 1 µl of dsDNase, and RNase-free ddH2O. The mixture was briefly centrifuged and then incubated at 37 °C for 2 min, followed by incubation at 55 °C for 10 min and at 85 °C for 10 s. The reaction product was immediately stored at -20 °C for subsequent experiments.
RT-qPCR analysis
The cDNA templates were then subjected to qPCR using SYBR Green Master (Roche, Switzerland). The following primers were designed by primer 5 and synthesized by GenePharma(China):SNHG16 forward primer: 5’-AATCGCCATGCGTTCTTTGG-3’; SNHG16 reverse primer: 5’-CAATCCTTGCAGTCCCATCG-3’; 18 S forward primer: 5’-GTAACCCGTTGAACCCCATT-3’; 18 S reverse primer: 5′CCATCCAATCGGTAGTAGCG–3’; 18 S was used as a reference gene. The 2 − ΔΔCt method was used to calculate the relative expression level. Experiments were repeated 3 times.
Urine cytology
Urine samples were collected before cystoscopic examination and any other treatments, and centrifuged at 1300 g for 10 min. The sediments were used for cytological analysis, and the diagnosis was confirmed by two cytopathologists.
Transmission electron microscopy (TEM)
Transmission electron microscopyIsolated exosomes were first resuspended in PBS, and then a 15 µL aliquot was absorbed onto carbon-coated Cu grids for 1 min.Subsequently, the grids were dyed using 15 µ L of 2.0% uranyl acetate for 1 min and allowed to dry for 15 min. The morphologyof isolated exosomes was identified by transmission electron microscopy (TEM; G2 spititi FEI; Tecnai).
Nanoparticle tracking analysis(NTA)
The size distribution of exosomes was determined usinga Delsa Nano Analyzer (DelsaNano, Beckman Coulter,Brea, CA, USA). The capture settings and analysis settings were performed manually according to the manufacturer’s instructions.
Western blots
Protein were prepared with a detergent buffer, and the protein concentration was determined using the bicinchoninic acid (BCA) protein assay (Solarbio, Beijin, China). Equal amounts of protein samples were separated by a 12% gel using sodium dodecyl sulfate-polyacrylamide gel electrophoresis(SDS-PAGE) and transferred onto PVDF membranes (Millipore, Billerica, MA, USA). The membranes were probed with anti-TSG101 (GeneTex, USA), anti-CD63 (Immunoway,USA) antibodies overnight at 4˚C. Immune complexes were detected by enhanced chemiluminescence (Proteintech, Chicago,USA).
Statistical analysis
All statistical analyses, ROC curve plotting and graphing were performed using SPSS 17.0 (IBM, SPSS, Chicago, IL, USA) and GraphPad Prism 5.0 (GraphPad Software, La Jolla, CA, USA). The Kolmogorov‒Smirnov test was used to analyse the distribution of each group of samples. Since the data did not follow a normal distribution, they are presented as the median (interquartile range). The nonparametric Mann‒Whitney U test was used to compare the expression levels of lncRNAs between the two groups. A two-sided P value of less than 0.05 was considered statistically significant.
Results
Identification of urinary exosomes in bladder cancer
TEM showed exosomes have a diameter of 30–150 nm with a cup-shaped membrane (Fig. 1a). NTA detected exosome particle size distribution, and the results showed that the highest particle size distribution peak in the exosome suspension was 123 nm (Fig. 1b). Western blot analysis of exosome marker proteins showed clear CD63 and TSG101 protein bands, but no protein expression was found in the exosome-depleted supernatant(EDS) (Fig. 1c). All the above results suggested that the enriched extracellular vesicles had the characteristics of exosomes.
Differential Analysis of lncRNA SNHG16 expression in urine exosomes between the two groups
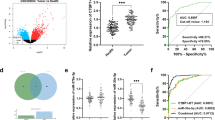
To determine whether there is a difference in the expression of lncRNA SNHG16 in urine exosomes between patients with bladder cancer and healthy controls, we first used RT‒qPCR to compare the differences in lncRNA SNHG16 expression between the bladder cancer cell line T24 and the benign urinary tract epithelial cell line SV-HUC-1, as well as between exosomes derived from these two cell sources. The results show that the expression of lncRNA SNHG16 was higher in T24 cells than in SV-HUC-1 cells (Fig. 2a) and higher in exosomes derived from T24 cells than in exosomes derived from SV-HUC-1 cells (Fig. 2b). Since bladder tumours originate from the urinary tract epithelium and the tumour is directly exposed to urine, we hypothesize that exosomes secreted by the tumour with high expression of lncRNA SNHG16 will directly enter urine. Therefore, the expression of lncRNA SNHG16 in urine exosomes of bladder cancer patients may be higher than that of healthy controls. Next, we used RT‒qPCR to detect the relative expression levels of urine exosomal lncRNA SNHG16 in the two groups. The results show that the expression levels of lncRNA SNHG16 in urine exosomes of the bladder cancer group and control group were 3.49 (2.17, 4.77) and 1.54 (0.86, 2.49), respectively, and the difference was statistically significant (P < 0.001, Fig. 2c), which means that urine exosomal lncRNA SNHG16 may become a potential diagnostic biomarker for bladder cancer. Finally, to investigate whether the expression of lncRNA SNHG16 in exosomes was stable, we performed an experiment using RT‒qPCR. The enriched urine exosome suspension was divided into three groups: the first group was the negative control group, the second group was treated with 5 µg RNaseA, and the third group was treated with 5% Triton X-100 and 5 µg RNaseA. The results show that compared with the control group, the expression of lncRNA SNHG16 in the RNaseA group did not change significantly, possibly due to the protection of exosomes. However, in the RNaseA + 5% Triton X-100 group, Triton X-100 destroyed the exosome membrane structure, leading to RNA degradation by RNaseA and a sharp decrease in lncRNA SNHG16 expression (Fig. 2d). Therefore, we believe that lncRNA SNHG16 mainly exists stably inside the exosome vesicles, with exosome vesicles as a “protective umbrella”. Therefore, urine exosomal lncRNA SNHG16 may be a stable diagnostic biomarker for bladder cancer.
Differential expression analysis of urinary exosomal lncRNA SNHG16 in the two groups. The expression of lncRNA SNHG16 was higher in T24 cells than in SV-HUC-1 cells (a), higher in exosomes derived from T24 cells than in those derived from SV-HUC-1 cells (b), and higher in exosomes from bladder cancer patients than in the control group (c). RT‒qPCR showed that lncRNA SNHG16 was stably expressed in exosome vesicles (d). *, P value< 0.05
Relationship between urinary exosomal lncRNA SNHG16 levels and clinical pathological characteristics of bladder cancer patients
According to the different clinical pathological characteristics of the tumours, the bladder cancer patients were grouped by age, sex, tumour T stage, pathological grade, and maximum tumour diameter, and the expression levels of urinary exosomal lncRNA SNHG16 were compared and analysed for differences in these clinical pathological parameters. The results showed no significant differences among the groups (P > 0.05) (Table 1).
Diagnostic value of the urinary exosomal lncRNA SNHG16 expression level for bladder cancer
Currently, noninvasive detection methods used for bladder cancer diagnosis in clinical practice mostly involve urinary cytology. Therefore, we compared the results of urinary cytology in the two groups and used ROC curve analysis to evaluate the diagnostic efficacy of urinary cytology for bladder cancer, which showed an AUC of 0.597 (95% CI: 0.475-0.719, sensitivity = 21.40%, specificity = 97.6%, Fig. 3a). Further analysis showed that the AUC of urinary exosomal lncRNA SNHG16 for diagnosing bladder cancer was 0.791 (95% CI: 0.695‐0.887, sensitivity = 61.90%, specificity = 83.3%, Fig. 3b), which was significantly better than that of urinary cytology.
Discussion
lncRNAs can serve as potential diagnostic, metastatic, and prognostic markers for bladder cancer. For example, the expression of lncRNAs such as TUC338 and PVT1 can be used as biomarkers for early diagnosis of bladder cancer [28, 29], while the expression of PCAT6, NRON, GAS6-AS2, SNHG3, lncRNA TP73-AS1, and LINC00641 can predict poor prognosis of bladder cancer [30,31,32,33,34,35]. In addition, lncRNAs can also serve as potential biomarkers for bladder cancer metastasis, such as DLX6-AS1, which is upregulated in bladder cancer tissue and cell lines and is associated with TNM stage progression, lymph node metastasis, and distant metastasis [36]. lncRNA SNHG16 is located on chromosome 17q25.1 and was first identified as an oncogene of neuroblastoma [37]. Cao [38] confirmed that the expression of lncRNA SNHG16 is significantly higher in bladder cancer tissue than in adjacent normal tissue and is associated with tumour metastasis, pathological staging, and overall survival of patients. Inhibition of SNHG16 expression can significantly suppress the proliferation, migration, and invasion of bladder cancer cells, and promote cell apoptosis, possibly through inhibition of the Wnt/β-catenin pathway [39].
LncRNA is susceptible to degradation by ribonucleases in bodily fluids and is typically found free-floating throughout the body through exosome transport. Exosome-carried lncRNAs have many advantages as tumour markers, including: (1) good specificity, sensitivity, and noninvasiveness; (2) noncoding RNAs packaged in exosomes can escape degradation by ribonucleases, making them stable; and (3) convenient sampling, with blood and urine being the most common sources of exosomes. Studies have reported that the expression of exosome-lncRNA H19 in serum of bladder cancer patients is upregulated [16], and exosome-lncRNA PTENP1 in plasma is downregulated [40], both of which can serve as good tumour markers for bladder cancer diagnosis. However, obtaining urine samples is more convenient and less painful for patients compared to blood samples. Research has shown that bladder cancer cells can secrete exosomes into urine, and stable lncRNA has been found in exosomes [41]. Zhan et al. [19] proposed a combination of lncRNAs derived from urine exosomes for bladder cancer diagnosis and recurrence prediction. The combination of three lncRNAs (MALAT1, PCAT-1, and SPRY4-IT1) had an area under the ROC curve (AUC) of 0.854 for bladder cancer diagnosis, which was significantly higher than that for urine cytology (0.619). In addition, the upregulation of PCAT-1 and MALAT1 was correlated with poor nonmuscle invasive bladder cancer-free survival. Another study showed that exosome-lncRNAs ANRIL and PCAT-1 in urine could serve as potential diagnostic biomarkers for bladder cancer, with AUCs of 0.7229 (sensitivity of 46.67% and specificity of 87.5%) and 0.7292 (sensitivity of 43.33% and specificity of 87.5%), respectively [42].
Currently, research has reported that the lncRNA SNHG16 is highly expressed in the serum exosomes of bladder cancer patients and can be used to differentiate between bladder cancer cases and healthy individuals [43]. This project aimed to investigate the feasibility of urinary exosomal SNHG16 as a diagnostic biomarker for bladder cancer. The results show that the expression of lncRNA SNHG16 was higher in T24 cells than in SV-HUC-1 cells, and its expression in exosomes derived from T24 cells was also higher than that from SV-HUC-1 cells. Since bladder cancer tumours are directly exposed to urine, it is inferred that exosomes with high expression of lncRNA SNHG16 secreted by bladder cancer may directly enter urine. Therefore, the expression of lncRNA SNHG16 in urinary exosomes of bladder cancer patients may be higher than that of healthy individuals. Further research showed that there was a statistically significant difference in the expression level of urinary exosomal lncRNA SNHG16 between the bladder cancer group and the control group (P < 0.001). This suggests that urinary exosomal lncRNA SNHG16 may be a noninvasive and stable diagnostic biomarker for bladder cancer. Finally, we used the relative expression value of RT‒qPCR to plot the ROC curve, and the results show that the diagnostic performance of urinary exosomal lncRNA SNHG16 for bladder cancer (AUC = 0.791, 95% CI: 0.695–0.887, sensitivity = 61.90%, specificity = 83.3%) was better than that of urinary cytology (AUC = 0.597, 95% CI: 0.475–0.719, sensitivity = 21.40%, specificity = 97.6%). In the data analysis, we did not find any correlation between the expression level of urinary exosomal lncRNA SNHG16 and age, sex, pathological T stage, pathological grade, or tumour size of bladder cancer patients (P > 0.05), which may require analysis of more samples to uncover hidden associations.
Conclusion
In summary, we believe that urinary exosomal lncRNA SNHG16 can serve as a noninvasive and reliable diagnostic biomarker for bladder cancer, but further large-scale and multicentre studies are still needed to discover whether the expression of urinary exosomal lncRNA SNHG16 is associated with the survival prognosis, pathological staging, and distant metastasis of bladder cancer patients.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AUC:
-
Transmission electron microscopy
- TEM:
-
Transmission electron microscopy
- NTA:
-
Nanoparticle tracking analysis
- ROC:
-
Receiver Operating Characteristic
- EDS:
-
Exosome-depleted supernatant
References
Babjuk M, Bohle A, Burger M, Capoun O, Cohen D, Comperat EM et al (2017) EAU Guidelines on Non-Muscle-invasive Urothelial Carcinoma of the bladder: Update 2016. Eur Urol 71(3):447–461
Aggen DH, Drake CG (2017) Biomarkers for immunotherapy in bladder cancer: a moving target. J Immunother Cancer 5(1):94
Kubota Y, Nakaigawa N (2016) Committee for Establishment of the Clinical Practice Guideline for the management of bladder C, the japanese Urological A. essential content of evidence-based clinical practice guidelines for bladder cancer: the japanese Urological Association 2015 update. Int J Urol 23(8):640–645
Clark PE, Spiess PE, Agarwal N, Bangs R, Boorjian SA, Buyyounouski MK et al (2016) NCCN Guidelines Insights: bladder Cancer, Version 2.2016. J Natl Compr Canc Netw 14(10):1213–1224
Giannopoulos A, Manousakas T, Mitropoulos D, Botsoli-Stergiou E, Constantinides C, Giannopoulou M et al (2000) Comparative evaluation of the BTAstat test, NMP22, and voided urine cytology in the detection of primary and recurrent bladder tumors. Urology 55(6):871–875
Gregoire M, Fradet Y, Meyer F, Tetu B, Bois R, Bedard G et al (1997) Diagnostic accuracy of urinary cytology, and deoxyribonucleic acid flow cytometry and cytology on bladder washings during followup for bladder tumors. J Urol 157(5):1660–1664
Grossman HB (1998) New methods for detection of bladder cancer. Semin Urol Oncol 16(1):17–22
Pode D, Shapiro A, Wald M, Nativ O, Laufer M, Kaver I (1999) Noninvasive detection of bladder cancer with the BTA stat test. J Urol 161(2):443–446
Ramakumar S, Bhuiyan J, Besse JA, Roberts SG, Wollan PC, Blute ML et al (1999) Comparison of screening methods in the detection of bladder cancer. J Urol 161(2):388–394
Zippe C, Pandrangi L, Potts JM, Kursh E, Novick A, Agarwal A (1999) NMP22: a sensitive, cost-effective test in patients at risk for bladder cancer. Anticancer Res 19(4A):2621–2623
Crozier J, Papa N, Perera M, Ngo B, Bolton D, Sengupta S et al (2019) Comparative sensitivity and specificity of imaging modalities in staging bladder cancer prior to radical cystectomy: a systematic review and meta-analysis. World J Urol 37(4):667–690
Schulz GB, Gresser EK, Casuscelli J, Strittmatter F, Tritschler S, Karl A et al (2019) [Value of imaging in upper urinary tract tumors]. Urologe A 58(1):5–13
Yang H, Fu H, Xu W, Zhang X (2016) Exosomal non-coding RNAs: a promising cancer biomarker. Clin Chem Lab Med 54(12):1871–1879
Li Q, Shao Y, Zhang X, Zheng T, Miao M, Qin L et al (2015) Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour Biol 36(3):2007–2012
Santangelo L, Giurato G, Cicchini C, Montaldo C, Mancone C, Tarallo R et al (2016) The RNA-Binding protein SYNCRIP is a component of the hepatocyte Exosomal Machinery Controlling MicroRNA sorting. Cell Rep 17(3):799–808
Wang J, Yang K, Yuan W, Gao Z (2018) Determination of serum exosomal H19 as a noninvasive biomarker for bladder Cancer diagnosis and prognosis. Med Sci Monit 24:9307–9316
Tao Y, Tang Y, Yang Z, Wu F, Wang L, Yang L et al (2020) Exploration of serum exosomal LncRNA TBILA and AGAP2-AS1 as promising biomarkers for diagnosis of Non-Small Cell Lung Cancer. Int J Biol Sci 16(3):471–482
Cai C, Zhang H, Zhu Y, Zheng P, Xu Y, Sun J et al (2019) Serum Exosomal Long Noncoding RNA pcsk2-2:1 as a potential Novel Diagnostic Biomarker for gastric Cancer. Onco Targets Ther 12:10035–10041
Zhan Y, Du L, Wang L, Jiang X, Zhang S, Li J et al (2018) Expression signatures of exosomal long non-coding RNAs in urine serve as novel non-invasive biomarkers for diagnosis and recurrence prediction of bladder cancer. Mol Cancer 17(1):142
Ding XZ, Zhang SQ, Deng XL, Qiang JH (2021) Serum exosomal lncRNA DLX6-AS1 is a Promising Biomarker for Prognosis Prediction of Cervical Cancer. Technol Cancer Res Treat 20:1533033821990060
Chen L, Qiu CH, Chen Y, Wang Y, Zhao JJ, Zhang M (2020) LncRNA SNHG16 drives proliferation, migration, and invasion of lung cancer cell through modulation of miR-520/VEGF axis. Eur Rev Med Pharmacol Sci 24(18):9522–9531
Du SM (2020) The SNHG16/miR-30a axis promotes breast cancer cell proliferation and invasion by regulating RRM2. Neoplasma 67(3):567–575
Ke D, Wang Q, Ke S, Zou L, Wang Q, Long-Non Coding (2020) RNA SNHG16 supports Colon cancer cell growth by modulating miR-302a-3p/AKT Axis. Pathol Oncol Res 26(3):1605–1613
Tao L, Wang X, Zhou Q (2020) Long noncoding RNA SNHG16 promotes the tumorigenicity of cervical cancer cells by recruiting transcriptional factor SPI1 to upregulate PARP9. Cell Biol Int 44(3):773–784
Xia W, Liu Y, Cheng T, Xu T, Dong M, Hu X (2021) Extracellular vesicles carry lncRNA SNHG16 to promote metastasis of breast Cancer cells via the miR-892b/PPAPDC1A Axis. Front Cell Dev Biol 9:628573
Yu L, Chen D, Song J (2020) LncRNA SNHG16 promotes non-small cell lung cancer development through regulating EphA2 expression by sponging miR-520a-3p. Thorac Cancer 11(3):603–611
Xiang Z, Huang G, Wu H, He Q, Yang C, Dou R et al (2022) SNHG16 upregulation-induced positive feedback loop with YAP1/TEAD1 complex in Colorectal Cancer cell lines facilitates liver metastasis of colorectal cancer by modulating CTCs epithelial-mesenchymal transition. Int J Biol Sci 18(14):5291–5308
Li G, Zhang Y, Mao J, Hu P, Chen Q, Ding W et al (2019) lncRNA TUC338 is a potential diagnostic biomarker for bladder cancer. J Cell Biochem 120(10):18014–18019
Yu C, Longfei L, Long W, Feng Z, Chen J, Chao L et al (2019) LncRNA PVT1 regulates VEGFC through inhibiting miR-128 in bladder cancer cells. J Cell Physiol 234(2):1346–1353
Zhang D, Du D, Yi S, Li X (2020) LncRNA PCAT6: a potential biomarker for diagnosis and prognosis of bladder cancer. Ann Diagn Pathol 49:151642
Xiong T, Huang C, Li J, Yu S, Chen F, Zhang Z et al (2020) LncRNA NRON promotes the proliferation, metastasis and EMT process in bladder cancer. J Cancer 11(7):1751–1760
Rui X, Wang L, Pan H, Gu T, Shao S, Leng J (2019) LncRNA GAS6-AS2 promotes bladder cancer proliferation and metastasis via GAS6-AS2/miR-298/CDK9 axis. J Cell Mol Med 23(2):865–876
Dai G, Huang C, Yang J, Jin L, Fu K, Yuan F et al (2020) LncRNA SNHG3 promotes bladder cancer proliferation and metastasis through miR-515-5p/GINS2 axis. J Cell Mol Med 24(16):9231–9243
Tuo Z, Zhang J, Xue W (2018) LncRNA TP73-AS1 predicts the prognosis of bladder cancer patients and functions as a suppressor for bladder cancer by EMT pathway. Biochem Biophys Res Commun 499(4):875–881
Li Z, Hong S, Liu Z (2018) LncRNA LINC00641 predicts prognosis and inhibits bladder cancer progression through miR-197-3p/KLF10/PTEN/PI3K/AKT cascade. Biochem Biophys Res Commun 503(3):1825–1829
Guo J, Chen Z, Jiang H, Yu Z, Peng J, Xie J et al (2019) The lncRNA DLX6-AS1 promoted cell proliferation, invasion, migration and epithelial-to-mesenchymal transition in bladder cancer via modulating Wnt/beta-catenin signaling pathway. Cancer Cell Int 19:312
Yu M, Ohira M, Li Y, Niizuma H, Oo ML, Zhu Y et al (2009) High expression of ncRAN, a novel non-coding RNA mapped to chromosome 17q25.1, is associated with poor prognosis in neuroblastoma. Int J Oncol 34(4):931–938
Cao X, Xu J, Yue D (2018) LncRNA-SNHG16 predicts poor prognosis and promotes tumor proliferation through epigenetically silencing p21 in bladder cancer. Cancer Gene Ther 25(1–2):10–17
Feng F, Chen A, Huang J, Xia Q, Chen Y, Jin X (2018) Long noncoding RNA SNHG16 contributes to the development of bladder cancer via regulating miR-98/STAT3/Wnt/beta-catenin pathway axis. J Cell Biochem 119(11):9408–9418
Zheng R, Du M, Wang X, Xu W, Liang J, Wang W et al (2018) Exosome-transmitted long non-coding RNA PTENP1 suppresses bladder cancer progression. Mol Cancer 17(1):143
Beckham CJ, Olsen J, Yin PN, Wu CH, Ting HJ, Hagen FK et al (2014) Bladder cancer exosomes contain EDIL-3/Del1 and facilitate cancer progression. J Urol 192(2):583–592
Abbastabar M, Sarfi M, Golestani A, Karimi A, Pourmand G, Khalili E (2020) Tumor-derived urinary exosomal long non-coding RNAs as diagnostic biomarkers for bladder cancer. EXCLI J 19:301–310
Zhang S, Du L, Wang L, Jiang X, Zhan Y, Li J et al (2019) Evaluation of serum exosomal LncRNA-based biomarker panel for diagnosis and recurrence prediction of bladder cancer. J Cell Mol Med 23(2):1396–1405
Acknowledgements
No applicable.
Funding
This study was supported by Science and Technology Project of Lu’an City in 2020 (NO.2020laskt04), Scientific Research Foundation of Anhui Medical University in 2021(NO. 2021xkj098), and the Health Research Project of Anhui Province in 2022 (NO.AHWJ2022c010 and NO.AHWJ2022a033).
Author information
Authors and Affiliations
Contributions
CYL and GYL designed all the research projects, analyzed data, and wrote the manuscript. PCX revised and proofread the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the ethics committees of The Second Hospital of Tianjin Medical University. Informed consent was obtained from all individual participants included in the study.
Consent for publication
Not applicable.
Competing interests
No financial competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, C., Xu, P., Shao, S. et al. The value of urinary exosomal lncRNA SNHG16 as a diagnostic biomarker for bladder cancer. Mol Biol Rep 50, 8297–8304 (2023). https://doi.org/10.1007/s11033-023-08667-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08667-z