Abstract
Background
SMAD4 is a potent tumor suppressor. SMAD4 loss increases genomic instability and plays a critical role in the DNA damage response that leads to skin cancer development. We aimed to investigate SMAD4 methylation effects on mRNA and protein expression of SMAD4 in cancer and healthy tissues from patients with basal cell carcinoma (BCC), cutaneous squamous cell carcinoma (cSCC), and basosquamous skin cancer (BSC).
Methods and results
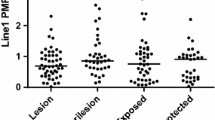
The study included 17 BCC, 24 cSCC and nine BSC patients. DNA and RNA were isolated from cancerous and healthy tissues following punch biopsy. Methylation-specific polymerase chain reaction (PCR) and real-time quantitative PCR methods were used to examine SMAD4 promoter methylation and SMAD4 mRNA levels, respectively. The percentage and intensity of staining of the SMAD4 protein were determined by immunohistochemistry. The percentage of SMAD4 methylation was increased in the patients with BCC (p = 0.007), cSCC (p = 0.004), and BSC (p = 0.018) compared to the healthy tissue. SMAD4 mRNA expression was decreased in the patients with BCC (p˂0.001), cSCC (p˂0.001), and BSC (p = 0.008). The staining characteristic of SMAD4 protein was negative in the cancer tissues of the patients with cSCC (p = 0.00). Lower SMAD4 mRNA levels were observed in the poorly differentiated cSCC patients (p = 0.001). The staining characteristics of the SMAD4 protein were related to age and chronic sun exposure.
Conclusions
Hypermethylation of SMAD4 and reduced SMAD4 mRNA expression were found to play a role in the pathogenesis of BCC, cSCC, and BSC. A decrease in SMAD4 protein expression level was observed only in cSCC patients. This suggests that epigenetic alterations to the SMAD4 gene are associated with cSCC.
Trial Registration
The name of the trial register: SMAD4 Methylation and Expression Levels in Non-melanocytic Skin Cancers; SMAD4 Protein Positivity.
The registration number: NCT04759261 (https://clinicaltrials.gov/ct2/results?term=NCT04759261).




Similar content being viewed by others
Data availability
All data generated or analysed during this study are included in this published article and its supplementary information files.
References
Ciążyńska M, Kamińska-Winciorek G, Lange D et al (2021) The incidence and clinical analysis of non-melanoma skin cancer. Sci Rep 11:4337. https://doi.org/10.1038/s41598-021-83502-8
Sung H, Ferlay J, Siegel RL et al (2021) Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71:209–249. https://doi.org/10.3322/caac.21660
Zambrano-Román M, Padilla-Gutiérrez JR, Valle Y, Muñoz-Valle JF, Valdés-Alvarado E (2022) Non-melanoma skin cancer: a genetic update and future perspectives. Cancers 14:2371. https://doi.org/10.3390/cancers14102371
Chiang A, Tan CZ, Kuonen F et al (2019) Genetic mutations underlying phenotypic plasticity in basosquamous carcinoma. J Invest Dermatol 139:2263-2271e5. https://doi.org/10.1016/j.jid.2019.03.1163
Volkenstein S, Wohlschlaeger J, Liebau J et al (2010) Basosquamous carcinoma–a rare but aggressive skin malignancy. J Plast Reconstr Aesthet Surg 63:e304–306. https://doi.org/10.1016/j.bjps.2009.05.058
Tarapore E, Atwood SX (2019) Defining the genetics of basosquamous carcinoma. J Invest Dermatol 139:2258–2260. https://doi.org/10.1016/j.jid.2019.04.011
Cives M, Mannavola F, Lospalluti L et al (2020) Non-melanoma skin cancers: biological and clinical features. Int J Mol Sci 21:5394. https://doi.org/10.3390/ijms21155394
Calzavara-Pinton P, Ortel B, Venturini M (2015) Non-melanoma skin cancer, sun exposure and sun protection. G Ital Dermatol Venereol 150:369–378
Hanahan D (2022) Hallmarks of cancer: new dimensions. Cancer Discov 12:31–46. https://doi.org/10.1158/2159-8290.Cd-21-1059
Pacella G, Capell BC (2021) Epigenetic and metabolic interplay in cutaneous squamous cell carcinoma. Exp Dermatol 30:1115–1125. https://doi.org/10.1111/exd.14354
Zhou C, Ye M, Ni S et al (2018) DNA methylation biomarkers for head and neck squamous cell carcinoma. Epigenetics 13:398–409. https://doi.org/10.1080/15592294.2018.1465790
Ke Y, Wang XJ (2021) TGFβ signaling in photoaging and UV-Induced skin cancer. J Invest Dermatol 141:1104–1110. https://doi.org/10.1016/j.jid.2020.11.007
Shao Y, Zhang J, Zhang R, Wan J, Zhang W, Yu B (2012) Examination of Smad2 and Smad4 copy-number variations in skin cancers. Clin Transl Oncol 14:138–142. https://doi.org/10.1007/s12094-012-0773-7
Kim Y, He YY (2014) Ultraviolet radiation-induced non-melanoma skin cancer: regulation of DNA damage repair and inflammation. Genes Dis 1:188–198. https://doi.org/10.1016/j.gendis.2014.08.005
Zhao M, Mishra L, Deng CX (2018) The role of TGF-β/SMAD4 signaling in cancer. Int J Biol Sci 14:111–123. https://doi.org/10.7150/ijbs.23230
Yang L, Mao C, Teng Y et al (2005) Targeted disruption of Smad4 in mouse epidermis results in failure of hair follicle cycling and formation of skin tumors. Cancer Res 65:8671–8678. https://doi.org/10.1158/0008-5472.Can-05-0800
Cai H, Sobue T, Kitamura T et al (2020) Epidemiology of nonmelanoma skin cancer in Japan: occupational type, lifestyle, and family history of cancer. Cancer Sci 111:4257–4265. https://doi.org/10.1111/cas.14619
Khalesi M, Whiteman DC, Rosendahl C et al (2015) Basal cell carcinomas on sun-protected vs. sun-exposed body sites: a comparison of phenotypic and environmental risk factors. Photodermatol Photoimmunol Photomed 31:202–211. https://doi.org/10.1111/phpp.12170
Reiter O, Mimouni I, Dusza S, Halpern AC, Leshem YA, Marghoob AA (2021) Dermoscopic features of basal cell carcinoma and its subtypes: a systematic review. J Am Acad Dermatol 85:653–664. https://doi.org/10.1016/j.jaad.2019.11.008
Sgouros D, Theofili M, Damaskou V et al (2021) Dermoscopy as a tool in differentiating cutaneous squamous cell carcinoma from its variants. Dermatol Pract Concept 11:e2021050. https://doi.org/10.5826/dpc.1102a50
Gambichler T, Skrygan M, Kaczmarczyk JM et al (2007) Increased expression of TGF-beta/Smad proteins in basal cell carcinoma. Eur J Med Res 12:509–514
Ozkan U, Ozcelik F, Yildiz M, Budak M (2019) Lipoprotein(a) gene polymorphism increases a risk factor for aortic valve calcification. J Cardiovasc Dev Dis 6:31. https://doi.org/10.3390/jcdd6030031
Gao T, Nie Y, Guo J (2012) Hypermethylation of the gene LARP2 for noninvasive prenatal diagnosis of β-thalassemia based on DNA methylation profile. Mol Biol Rep 39:6591–6598. https://doi.org/10.1007/s11033-012-1489-z
Guo W, Dong Z, Guo Y, Kuang G, Yang Z, Shan B (2012) Concordant repression and aberrant methylation of transforming growth factor-beta signaling pathway genes occurs early in gastric cardia adenocarcinoma. Mol Biol Rep 39:9453–9462. https://doi.org/10.1007/s11033-012-1810-x
Li LC, Dahiya R (2002) MethPrimer: designing primers for methylation PCRs. Bioinformatics 18:1427–1431. https://doi.org/10.1093/bioinformatics/18.11.1427
Rao X, Huang X, Zhou Z, Lin X (2013) An improvement of the 2ˆ(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinforma Biomath 3:71–85
Lin Z, Zhang L, Zhou J, Zheng J (2019) Silencing Smad4 attenuates sensitivity of colorectal cancer cells to cetuximab by promoting epithelialmesenchymal transition. Mol Med Rep 20:3735–3745. https://doi.org/10.3892/mmr.2019.10597
Xavier-Junior JCC, Ocanha-Xavier JP (2019) WHO,(2018) Classification of Skin Tumors. Am J Dermatopathol 41:699–700. https://doi.org/10.1097/dad.0000000000001446
Woo WA, Suarez MFB, Keohane SG (2021) Summary of the updated 2020 guidelines for cutaneous squamous cell carcinoma. Clin Exp Dermatol 46:1174–1177. https://doi.org/10.1111/ced.14587
He SM, Zhao ZW, Wang Y et al (2011) Reduced expression of SMAD4 in gliomas correlates with progression and survival of patients. J Exp Clin Cancer Res 30:70. https://doi.org/10.1186/1756-9966-30-70
Hernandez AL, Young CD, Wang JH, Wang XJ (2019) Lessons learned from SMAD4 loss in squamous cell carcinomas. Mol Carcinog 58:1648–1655. https://doi.org/10.1002/mc.23049
Bellizzi AM (2020) An algorithmic immunohistochemical approach to define tumor type and assign site of origin. Adv Anat Pathol 27:114–163. https://doi.org/10.1097/pap.0000000000000256
Hoot KE, Lighthall J, Han G et al (2008) Keratinocyte-specific Smad2 ablation results in increased epithelial-mesenchymal transition during skin cancer formation and progression. J Clin Invest 118:2722–2732. https://doi.org/10.1172/jci33713
Mitra D, Fernandez P, Bian L et al (2013) Smad4 loss in mouse keratinocytes leads to increased susceptibility to UV carcinogenesis with reduced Ercc1-mediated DNA repair. J Invest Dermatol 133:2609–2616. https://doi.org/10.1038/jid.2013.213
Hatziapostolou M, Iliopoulos D (2011) Epigenetic aberrations during oncogenesis. Cell Mol Life Sci 68:1681–1702. https://doi.org/10.1007/s00018-010-0624-z
Swellam M, Saad EA, Sabry S, Denewer A, Abdel Malak C, Abouzid A (2021) Alterations of PTEN and SMAD4 methylation in diagnosis of breast cancer: implications of methyl II PCR assay. J Genet Eng Biotechnol 19:54. https://doi.org/10.1186/s43141-021-00154-x
Yan W, Wistuba II, Emmert-Buck MR, Erickson HS (2011) Squamous cell carcinoma—similarities and differences among anatomical sites. Am J Cancer Res 1:275–300
Wu F, Weigel KJ, Zhou H, Wang XJ (2018) Paradoxical roles of TGF-β signaling in suppressing and promoting squamous cell carcinoma. Acta Biochim Biophys Sin (Shanghai) 50:98–105. https://doi.org/10.1093/abbs/gmx127
Bornstein S, White R, Malkoski S et al (2009) Smad4 loss in mice causes spontaneous head and neck cancer with increased genomic instability and inflammation. J Clin Invest 119:3408–3419. https://doi.org/10.1172/jci38854
Franzen A, Bootz F, Dietrich D (2020) Prognostic and predictive methylation biomarkers in HNSCC: Epigenomic diagnostics for head and neck squamous cell carcinoma (HNSCC). HNO 68:911–915. https://doi.org/10.1007/s00106-020-00902-4
Nikolouzakis TK, Falzone L, Lasithiotakis K et al (2020) Current and future trends in molecular biomarkers for diagnostic, prognostic, and predictive purposes in non-melanoma skin cancer. J Clin Med 9:2868. https://doi.org/10.3390/jcm9092868
Lin LH, Chang KW, Cheng HW, Liu CJ (2019) SMAD4 somatic mutations in head and neck carcinoma are associated with tumor progression. Front Oncol 9:1379. https://doi.org/10.3389/fonc.2019.01379
Hervás-Marín D, Higgins F, Sanmartín O et al (2019) Genome wide DNA methylation profiling identifies specific epigenetic features in high-risk cutaneous squamous cell carcinoma. PLoS ONE 14:e0223341. https://doi.org/10.1371/journal.pone.0223341
Heitzer E, Bambach I, Dandachi N, Horn M, Wolf P (2010) PTCH promoter methylation at low level in sporadic basal cell carcinoma analysed by three different approaches. Exp Dermatol 19:926–928. https://doi.org/10.1111/j.1600-0625.2010.01120.x
Brinkhuizen T, van den Hurk K, Winnepenninckx VJ et al (2012) Epigenetic changes in basal cell carcinoma affect SHH and WNT signaling components. PLoS ONE 7:e51710. https://doi.org/10.1371/journal.pone.0051710
Qiao W, Li AG, Owens P, Xu X, Wang XJ, Deng CX (2006) Hair follicle defects and squamous cell carcinoma formation in Smad4 conditional knockout mouse skin. Oncogene 25:207–217. https://doi.org/10.1038/sj.onc.1209029
Lange D, Persson U, Wollina U et al (1999) Expression of TGF-beta related smad proteins in human epithelial skin tumors. Int J Oncol 14:1049–1056. https://doi.org/10.3892/ijo.14.6.1049
Pietkiewicz P, Gornowicz-Porowska J, Bowszyc-Dmochowska M et al (2014) Discordant expression of desmoglein 2 and 3 at the mRNA and protein levels in nodular and superficial basal cell carcinoma revealed by immunohistochemistry and fluorescent in situ hybridization. Clin Exp Dermatol 39:628–635. https://doi.org/10.1111/ced.12355
Li JJ, Biggin MD (2015) Gene expression. Statistics requantitates the central dogma. Science 347:1066–1067. https://doi.org/10.1126/science.aaa8332
Fortelny N, Overall CM, Pavlidis P, Freue GVC (2017) Can we predict protein from mRNA levels? Nature 547:E19–E20. https://doi.org/10.1038/nature22293
Funding
This study was supported by the Trakya University Research Project Fund (2020/55).
Author information
Authors and Affiliations
Contributions
YGÜ and MB designed the research study. MB and EUK performed the research. YGÜ and MB carried out the data analysis and writing—review and editing. All the authors discussed the results and contributed to the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors agree that there are no conflicts of interest to declare.
Ethical approval
Approval for the study was granted by the Scientific Research Ethics Committee of the Trakya University Faculty of Medicine (approval number: 18/14, date: 06/11/2019).
Informed consent
All the subjects who agreed to participate in the study gave their written informed consent.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gürsel Ürün, Y., Budak, M. & Usturalı Keskin, E. Methylation status, mRNA and protein expression of the SMAD4 gene in patients with non-melanocytic skin cancers. Mol Biol Rep 50, 7295–7304 (2023). https://doi.org/10.1007/s11033-023-08656-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08656-2