Abstract
Background
Plant microRNA, often known as miRNA, is a novel form of gene expression regulator that is known to play a significant role in phosphate starvation. The identification of microRNAs involved in the response to phosphate starvation in oil palms is beneficial for breeding programs.
Method
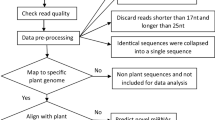
The main nursery stage seedlings of two oil palm progenies were treated with three different fertiliser namely: complete fertiliser with urea, P2O5, K2O, and MgO based on the standard procedure as a control (C); fertiliser with urea, K2O, MgO without P2O5 (P0); and no fertiliser (F0) for 24 weeks. A total of six oil palm roots were subjected to RNA isolation, followed by miRNA sequencing using the Illumina HiSeq 4000 platform, and all reads were computationally analysed.
Results
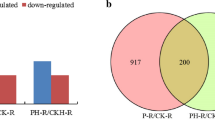
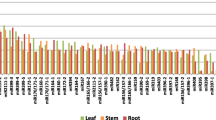
In total, 119 potential miRNAs related to 5,891 genes were identified. The P-specific miRNAs were assumed based on the miRNAs that identified without P fertilizer treatment, resulted of twenty miRNA sequences in the treatment comparison of (C vs P0) vs (C vs F0). Those 20 miRNA sequences were grouped into 9 families, namely EgmiR319; EgmiR399; EgmiR396; EgmiR172; EgmiR156; EgmiR157; miR5648; miR5645; and EgmiRNA_unidentified. Two miRNAs were selected for RT-qPCR validation, namely EgMir399 and EgMir172. Their expression pattern was similar with the RNA sequencing results and shown opposite expression pattern with their target genes, UBC E2 24 and APETALA2, respectively.
Conclusions
The nine micro RNA families was identified in oil palm root tissue at phosphate starvation.








Similar content being viewed by others
Data availability
The datasets supporting the conclusions of this article are included in this.
Abbreviations
- P:
-
Phosphate
- Pi:
-
Inorganic phosphate
- mRNA:
-
Messenger RNA
- miRNA:
-
MicroRNA
- C:
-
Complete fertiliser (dozage based on standard procedure)
- P0:
-
SOP fertiliser without phosphate
- F0:
-
No fertilizers
- RT-qPCR:
-
Reverse transcription-quantitative polymerase chain reaction
- DxP:
-
Oil palm progeny that from Dura and Pisifera crossing called Tenera
- Ho_P0:
-
Hoagland solution medium with 0% phosphorus
- Ho_P65:
-
Hoagland solution medium with 65% phosphorus
- ABMix:
-
Standard commercial nutrient solution
- cDNA:
-
Complementary DNA
- LncRNA:
-
Long non-coding RNA
- sRNA:
-
Small RNA
- rRNA:
-
Ribosome RNA
- GO:
-
Gene ontology
- BLAST:
-
Basic local alignment search tool
- RP:
-
Reverse primer
- FP:
-
Forward primers
- Ct:
-
Cycle thresholds
- MFE:
-
Minimum free energy
References
Kamerlin SCL, Sharma PK, Prasad RB, Warshel A (2013) Why nature really chose phosphate. Q Rev Biophys 46:1–132. https://doi.org/10.1017/S0033583512000157
Vance CP, Uhde-Stone C, Allan DL (2003) Phosphorus acquisition and use: Critical adaptations by plants for securing a nonrenewable resource. New Phytol 157:423–447. https://doi.org/10.1046/j.1469-8137.2003.00695.x
Tiemann TT, Donough CR, Lim YL et al (2018) Feeding the Palm: a review of oil palm nutrition. Adv Agron 152:149–243. https://doi.org/10.1016/bs.agron.2018.07.001
Mohidin H, Hanafi MM, Rafii YM et al (2015) Determination of optimum levels of nitrogen, phosphorus and potassium of oil palm seedlings in solution culture. Bragantia 74:247–254. https://doi.org/10.1590/1678-4499.0408
Rogers K, Chen X (2013) Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25:2383–2399. https://doi.org/10.1105/tpc.113.113159
Liang G, He H, Yu D (2012) Identification of nitrogen starvation-responsive microRNAs in Arabidopsis thaliana. PLoS ONE. https://doi.org/10.1371/journal.pone.0048951
Ren Y, Sun F, Hou J et al (2015) Differential profiling analysis of miRNAs reveals a regulatory role in low N stress response of Populus. Funct Integr Genomics 15:93–105. https://doi.org/10.1007/s10142-014-0408-x
Xu F, Liu Q, Chen L et al (2013) Genome-wide identification of soybean microRNAs and their targets reveals their organ-specificity and responses to phosphate starvation. BMC Genom. https://doi.org/10.1186/1471-2164-14-66
Fujii H, Chiou TJ, Lin SI et al (2005) A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol 15:2038–2043. https://doi.org/10.1016/j.cub.2005.10.016
Hackenberg M, Shi BJ, Gustafson P, Langridge P (2013) Characterization of phosphorus-regulated miR399 and miR827 and their isomirs in barley under phosphorus-sufficient and phosphorus-deficient conditions. BMC Plant Biol. https://doi.org/10.1186/1471-2229-13-214
Paul S, Datta SK, Datta K (2015) miRNA regulation of nutrient homeostasis in plants. Front Plant Sci 6:1–11. https://doi.org/10.3389/fpls.2015.00232
Lei KJ, Lin YM, An GY (2016) miR156 modulates rhizosphere acidification in response to phosphate limitation in Arabidopsis. J Plant Res 129:275–284. https://doi.org/10.1007/s10265-015-0778-8
Kumar S, Verma S, Trivedi PK (2017) Involvement of small RNAs in phosphorus and sulfur sensing, signaling and stress: current update. Front Plant Sci 8:1–12. https://doi.org/10.3389/fpls.2017.00285
Chiou T, Aung K, Lin S et al (2006) Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 18:412–421. https://doi.org/10.1105/tpc.105.038943.1
Aung K, Lin SI, Wu CC et al (2006) pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol 141:1000–1011. https://doi.org/10.1104/pp.106.078063
Bari R, Pant BD, Stitt M, Scheible WR (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141:988–999. https://doi.org/10.1104/pp.106.079707
Li Z, Zhang X, Liu X et al (2016) miRNA alterations are important mechanism in maize adaptations to low-phosphate environments. Plant Sci 252:103–117. https://doi.org/10.1016/j.plantsci.2016.07.009
Lundmark M, Kørner CJ, Nielsen TH (2010) Global analysis of microRNA in Arabidopsis in response to phosphate starvation as studied by locked nucleic acid-based microarrays. Physiol Plant 140:57–68. https://doi.org/10.1111/j.1399-3054.2010.01384.x
Zeng HQ, Zhu YY, Huang SQ, Yang ZM (2010) Analysis of phosphorus-deficient responsive miRNAs and cis-elements from soybean (Glycine max L.). J Plant Physiol 167:1289–1297. https://doi.org/10.1016/j.jplph.2010.04.017
Nasaruddin N, Harikrishna K, Othman RY et al (2007) Computational prediction of microRNAs from Oil Palm (Elaeis guineensis Jacq.) expressed sequence tags. Genetics 15:107–113
Ho H, Gudimella R, Ong-Abdullah M, Harikrishna JA (2017) Expression of microRNAs during female inflorescence development in African oil palm (Elaeis guineensis Jacq.). Tree Genet Genomes 13:35. https://doi.org/10.1007/s11295-017-1120-5
Friedländer MR, MacKowiak SD, Li N et al (2012) MiRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res 40:37–52. https://doi.org/10.1093/nar/gkr688
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:1–21. https://doi.org/10.1186/s13059-014-0550-8
Bonnet E, He Y, Billiau K, van de Peer Y (2010) TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics 26:1566–1568. https://doi.org/10.1093/bioinformatics/btq233
Shannon Markiel A, Owen Ozier NS, Baliga JT, Wang DR et al (2003) Cytoscape: a software environment for integrated models. Genome Res 13:426. https://doi.org/10.1101/gr.1239303.metabolite
Kozomara A, Birgaoanu M, Griffiths-Jones S (2019) MiRBase: from microRNA sequences to function. Nucleic Acids Res 47:D155–D162. https://doi.org/10.1093/nar/gky1141
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Hofacker IL, Fontana W, Stadler PF et al (1994) Fast folding and comparison of RNA secondary structures. Monatshefte für Chemie Chem Mon 125:167–188. https://doi.org/10.1007/BF00818163
Chien PS, Bin CC, Wang Z, Chiou TJ (2017) MicroRNA-mediated signaling and regulation of nutrient transport and utilization. Curr Opin Plant Biol 39:73–79. https://doi.org/10.1016/j.pbi.2017.06.007
Khraiwesha et al (2012) Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim Biophys Acta BBA - Gene Regul Mech 1819:137–148. https://doi.org/10.1016/j.bbagrm.2011.05.001
Pant BD, Buhtz A, Kehr J, Scheible WR (2008) MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J 53:731–738. https://doi.org/10.1111/j.1365-313X.2007.03363.x
Kruszka K, Pieczynski M, Windels D et al (2012) Role of microRNAs and other sRNAs of plants in their changing environments. J Plant Physiol 169:1664–1672. https://doi.org/10.1016/j.jplph.2012.03.009
Zhang J, Lin Y, Wu F et al (2021) Profiling of microRNAs and their targets in roots and shoots reveals a potential MiRNA-mediated interaction network in response to phosphate deficiency in the forestry tree betula luminifera. Front Genet 12:1–17. https://doi.org/10.3389/fgene.2021.552454
Lei KJ, Lin YM, Ren J et al (2016) Modulation of the phosphate-deficient responses by MicroRNA156 and its targeted SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 3 in arabidopsis. Plant Cell Physiol 57:192–203. https://doi.org/10.1093/pcp/pcv197
Baek D, Kim MC, Chun HJ et al (2013) Regulation of miR399f transcription by AtMYB2 affects phosphate starvation responses in Arabidopsis. Plant Physiol 161:362–373. https://doi.org/10.1104/pp.112.205922
Franco-Zorrilla JM, Valli A, Todesco M et al (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39:1033–1037. https://doi.org/10.1038/ng2079
Zeng H, Wang G, Hu X et al (2014) Role of microRNAs in plant responses to nutrient stress. Plant Soil 374:1005–1021. https://doi.org/10.1007/s11104-013-1907-6
Yan Z, Hossain MS, Wang J et al (2013) miR172 regulates soybean nodulation. Mol Plant-Microbe Interact 26:1371–1377. https://doi.org/10.1094/MPMI-04-13-0111-R
Wang Y, Wang L, Zou Y et al (2014) Soybean miR172c targets the repressive AP2 transcription factor NNC1 to activate ENOD40 expression and regulate nodule initiation. Plant Cell 26:4782–4801. https://doi.org/10.1105/tpc.114.131607
Bhogale S, Mahajan AS, Natarajan B et al (2014) MicroRNA156: A potential graft-transmissible microrna that modulates plant architecture and tuberization in Solanum tuberosum ssp. andigena. Plant Physiol 164:1011–1027. https://doi.org/10.1104/pp.113.230714
Xu M, Hu T, Zhao J et al (2016) Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genet 12:1–29. https://doi.org/10.1371/journal.pgen.1006263
Yu N, Niu QW, Ng KH, Chua NH (2015) The role of miR156/SPLs modules in Arabidopsis lateral root development. Plant J 83:673–685. https://doi.org/10.1111/tpj.12919
Nag A, King S, Jack T (2009) miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc Natl Acad Sci USA 106:22534–22539. https://doi.org/10.1073/pnas.0908718106
Muneer S, Jeong BR (2015) Proteomic analysis provides new insights in phosphorus homeostasis subjected to pi (inorganic phosphate) starvation in tomato plants (Solanum lycopersicum L.). PLoS ONE 10:1–18. https://doi.org/10.1371/journal.pone.0134103
Zhou M, Li D, Li Z et al (2013) Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiol 161:1375–1391. https://doi.org/10.1104/pp.112.208702
Lu YT, Li MY, Cheng KT et al (2014) Transgenic plants that express the phytoplasma effector SAP11 show altered phosphate starvation and defense responses. Plant Physiol 164:1456–1469. https://doi.org/10.1104/pp.113.229740
Ling LZ, Zhang SD, Zhao F et al (2017) Transcriptome-wide identification and prediction of MiRNAS and their targets in Paris polyphylla var. Yunnanensis by high-throughput sequencing analysis. Int J Mol Sci 18:1–12. https://doi.org/10.3390/ijms18010219
Kuo H-F, Chiou T-J (2011) The role of MicroRNAs in phosphorus deficiency signaling. Plant Physiol 156:1016–1024. https://doi.org/10.1104/pp.111.175265
Aung K, Lin S, Wu C et al (2006) pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a MicroRNA399 target gene 1. Plant Physiol 141:1000–1011. https://doi.org/10.1104/pp.106.078063.solution
Nova-Franco B, Íñiguez LP, Valdés-López O et al (2015) The micro-RNA72c-APETALA2-1 node as a key regulator of the common bean-Rhizobium etli nitrogen fixation symbiosis. Plant Physiol 168:273–291. https://doi.org/10.1104/pp.114.255547
Chen Y, Wu P, Zhao Q et al (2018) Overexpression of a phosphate starvation response ap2/erf gene from physic nut in arabidopsis alters root morphological traits and phosphate starvation-induced anthocyanin accumulation. Front Plant Sci 9:1–12. https://doi.org/10.3389/fpls.2018.01186
Kong S, Nor S, Abdullah A et al (2021) Comparative transcriptome analysis reveals novel insights into transcriptional responses to phosphorus starvation in oil palm (Elaeis guineensis) root. BMC Genom Data. https://doi.org/10.1186/s12863-021-00962-7
Acknowledgements
We thank Ogi Ajitio Ramadhan and Redy Wahyu Permana for their technical support. We thank Reno Tryono, Redi Aditama, and Chris Darmawan, who read and improved the manuscript.
Funding
This work was funded by PT SMART Tbk under a research project code 3.1.1.057.
Author information
Authors and Affiliations
Contributions
TIS, RBD, and SDM: conceived and designed the experiments. TIS and SDM: performed the experiments. TIS and ZAT: analyzed the data. TIS and RBD: wrote the paper. CU and TL: funding acquisition, review and supervision. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
Not applicable.
Informed consent
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Saputra, T.I., Roberdi, Maryanto, S.D. et al. Identification of microRNAs involved in the Phosphate starvation response in Oil Palm (Elaeis guineensis Jacq.). Mol Biol Rep 50, 5609–5620 (2023). https://doi.org/10.1007/s11033-023-08484-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08484-4