Abstract
Background
Concerning the different clinical behavior of epidermal growth factor receptor (EGFR) subtypes in advanced-stage lung cancer patients, the current study aimed to evaluate the clinical, pathological, and prognostic significance of EGFR mutation subtypes, and treatment response in patients with advanced-stage lung cancer.
Methods and results
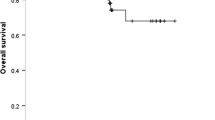
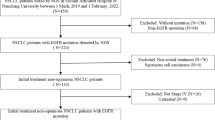
A retrospective study enrolled a total of 346 patients with advanced-stage lung cancer tested for EGFR mutation. EGFR mutation was analyzed by amplification refractory mutation system-polymerase chain reaction (ARMS-PCR). Statistical analysis was performed using SPSS version 20.0. EGFR mutation was evident in 38% of patients with the highest prevalence of exon 19 deletions. A higher incidence of 19-deletions and 20-insertions were observed in young patients, while a higher incidence of L858R was noted in old age patients. Patients with de-novo T790M failed to improve their OS by any of the treatment modalities. Patients with de-novo T790M mutation have a higher risk of developing lung, liver, and multiple site metastases while patients with L858R mutation have a higher risk of developing brain metastasis. Additionally, patients with 19 deletion mutation did not improve their OS after receiving conventional chemotherapy hence, they demonstrate better survival only after EGFR-TKIs. Multivariate survival analysis predicted chemotherapy as an independent predictor of OS.
Conclusion
Besides clinicopathological and prognostic consequences of EGFR mutation and mutation subtypes, patients harboring TKI sensitive, or insensitive mutations reveal different secondary disease development and hence should be treated accordingly for better survival. Current findings may provide the basis for a better treatment strategy.





Similar content being viewed by others
Data Availability
All data generated or analysed during this study are included in this published article.
References
Sung H, Ferlay J, Siegel RL et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71(3):209–249. https://doi.org/10.3322/caac.21660
Siegel R, Naishadham D, Jemal A (2013) Cancer statistics, 2013. CA Cancer J Clin 63(1):11–30. https://doi.org/10.3322/caac.21166
Chen Z, Fillmore CM, Hammerman PS et al (2014) Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer 14(8):535–546. https://doi.org/10.1038/nrc3775
Tsao MS, Sakurada A, Cutz JC et al (2005) Erlotinib in lung cancer - molecular and clinical predictors of outcome. N Engl J Med 353(2):133–144. https://doi.org/10.1056/NEJMoa050736
Shaw AT, Kim DW, Nakagawa K et al (2013) Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 368(25):2385–2394. https://doi.org/10.1056/NEJMoa1214886
Kris MG, Johnson BE, Berry LD et al (2014) Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 311(19):1998–2006. https://doi.org/10.1001/jama.2014.3741
Yoshida T, Zhang G, Haura EB (2010) Targeting epidermal growth factor receptor: central signaling kinase in lung cancer. Biochem Pharmacol 80(5):613–623. https://doi.org/10.1016/j.bcp.2010.05.014
Janne PA, Engelman JA, Johnson BE (2005) Epidermal growth factor receptor mutations in non-small-cell lung cancer: implications for treatment and tumor biology. J Clin Oncol 23(14):3227–3234. https://doi.org/10.1200/JCO.2005.09.985
Wang Y, Deng G, Liu X et al (2013) Monoclonal antibodies in lung cancer. Expert Opin Biol Ther 13(2):209–226. https://doi.org/10.1517/14712598.2012.748742
Perez R, Crombet T, de Leon J et al (2013) A view on EGFR-targeted therapies from the oncogene-addiction perspective. Front Pharmacol 4:53. https://doi.org/10.3389/fphar.2013.00053
Remon J, Moran T, Majem M et al (2014) Acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in EGFR-mutant non-small cell lung cancer: a new era begins. Cancer Treat Rev 40(1):93–101. https://doi.org/10.1016/j.ctrv.2013.06.002
Yu HA, Riely GJ, Lovly CM (2014) Therapeutic strategies utilized in the setting of acquired resistance to EGFR tyrosine kinase inhibitors. Clin Cancer Res 20(23):5898–5907. https://doi.org/10.1158/1078-0432.CCR-13-2437
Lynch TJ, Bell DW, Sordella R et al (2004) Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 350(21):2129–2139. https://doi.org/10.1056/NEJMoa040938
Paez JG, Janne PA, Lee JC et al (2004) EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304(5676):1497–1500. https://doi.org/10.1126/science.1099314
Kobayashi Y, Mitsudomi T (2016) Not all epidermal growth factor receptor mutations in lung cancer are created equal: perspectives for individualized treatment strategy. Cancer Sci 107(9):1179–1186. https://doi.org/10.1111/cas.12996
Mok TS, Wu YL, Thongprasert S et al (2009) Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 361(10):947–957. https://doi.org/10.1056/NEJMoa0810699
Wu YL, Zhou C, Hu CP et al (2014) Afatinib versus cisplatin plus gemcitabine for first-line treatment of asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol 15(2):213–222. https://doi.org/10.1016/S1470-2045(13)70604-1
Lin Y, Wang X, Jin H (2014) EGFR-TKI resistance in NSCLC patients: mechanisms and strategies. Am J Cancer Res 4(5):411–435
Riely GJ, Yu HA (2015) EGFR: the paradigm of an Oncogene-Driven lung cancer. Clin Cancer Res 21(10):2221–2226. https://doi.org/10.1158/1078-0432.CCR-14-3154
Joshi J, Raval A, Desai U et al (2021) EGFR Mutation Analysis in Non-small Cell Lung Carcinoma Patients: a Liquid Biopsy Approach. Indian J Clin Biochem 36(1):51–58. https://doi.org/10.1007/s12291-019-00864-7
Kim HJ, Oh SY, Kim WS et al (2013) Clinical investigation of EGFR mutation detection by pyrosequencing in lung cancer patients. Oncol Lett 5(1):271–276. https://doi.org/10.3892/ol.2012.950
Dufort S, Richard MJ, de Fraipont F (2009) Pyrosequencing method to detect KRAS mutation in formalin-fixed and paraffin-embedded tumor tissues. Anal Biochem 391(2):166–168. https://doi.org/10.1016/j.ab.2009.05.027
Tsiatis AC, Norris-Kirby A, Rich RG et al (2010) Comparison of Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of KRAS mutations: diagnostic and clinical implications. J Mol Diagn 12(4):425–432. https://doi.org/10.2353/jmoldx.2010.090188
Ettinger DS, Akerley W, Bepler G et al (2010) Non-small cell lung cancer. J Natl Compr Canc Netw 8(7):740–801. https://doi.org/10.6004/jnccn.2010.0056
Li D, Ding L, Ran W et al (2020) Status of 10 targeted genes of non-small cell lung cancer in eastern China: a study of 884 patients based on NGS in a single institution. Thorac Cancer 11(9):2580–2589. https://doi.org/10.1111/1759-7714.13577
Zhao M, Zhan C, Li M et al (2018) Aberrant status and clinicopathologic characteristic associations of 11 target genes in 1,321 chinese patients with lung adenocarcinoma. J Thorac Dis 10(1):398–407. https://doi.org/10.21037/jtd.2017.12.68
Li S, Li L, Zhu Y et al (2014) Coexistence of EGFR with KRAS, or BRAF, or PIK3CA somatic mutations in lung cancer: a comprehensive mutation profiling from 5125 chinese cohorts. Br J Cancer 110(11):2812–2820. https://doi.org/10.1038/bjc.2014.210
Liu R, Zhou J, Ling X (2022) Optimizing patient outcomes through sequential EGFR TKI treatment in asian patients with EGFR mutation-positive NSCLC. Clin Med Insights Oncol 16:11795549221103215. https://doi.org/10.1177/11795549221103215
Suda K, Mitsudomi T, Shintani Y et al (2021) Clinical impacts of EGFR Mutation Status: analysis of 5780 surgically resected Lung Cancer cases. Ann Thorac Surg 111(1):269–276. https://doi.org/10.1016/j.athoracsur.2020.05.041
Toh CK, Gao F, Lim WT et al (2006) Never-smokers with lung cancer: epidemiologic evidence of a distinct disease entity. J Clin Oncol 24(15):2245–2251. https://doi.org/10.1200/JCO.2005.04.8033
Jin Y, Chen M, Yu X (2016) Differences among lesions with exon 19, exon 21 EGFR mutations and wild types in surgically resected non-small cell lung cancer. Sci Rep 6:31636. https://doi.org/10.1038/srep31636
Ohba T, Toyokawa G, Kometani T et al (2014) Mutations of the EGFR and K-ras genes in resected stage I lung adenocarcinoma and their clinical significance. Surg Today 44(3):478–486. https://doi.org/10.1007/s00595-013-0589-2
Ohba T, Toyokawa G, Osoegawa A et al (2016) Mutations of the EGFR, K-ras, EML4-ALK, and BRAF genes in resected pathological stage I lung adenocarcinoma. Surg Today 46(9):1091–1098. https://doi.org/10.1007/s00595-015-1295-z
Izar B, Sequist L, Lee M et al (2013) The impact of EGFR mutation status on outcomes in patients with resected stage I non-small cell lung cancers. Ann Thorac Surg 96(3):962–968. https://doi.org/10.1016/j.athoracsur.2013.05.091
Yang CY, Lin MW, Chang YL et al (2014) Programmed cell death-ligand 1 expression in surgically resected stage I pulmonary adenocarcinoma and its correlation with driver mutations and clinical outcomes. Eur J Cancer 50(7):1361–1369. https://doi.org/10.1016/j.ejca.2014.01.018
Nishii T, Yokose T, Miyagi Y et al (2017) Prognostic value of EGFR mutations in surgically resected pathological stage I lung adenocarcinoma. Asia Pac J Clin Oncol 13(5):e204–e211. https://doi.org/10.1111/ajco.12512
Liu WS, Zhao LJ, Pang QS et al (2014) Prognostic value of epidermal growth factor receptor mutations in resected lung adenocarcinomas. Med Oncol 31(1):771. https://doi.org/10.1007/s12032-013-0771-9
Lin CY, Wu YM, Hsieh MH et al (2017) Prognostic implication of EGFR gene mutations and histological classification in patients with resected stage I lung adenocarcinoma. PLoS ONE 12(10):e0186567. https://doi.org/10.1371/journal.pone.0186567
D’Angelo SP, Janjigian YY, Ahye N et al (2012) Distinct clinical course of EGFR-mutant resected lung cancers: results of testing of 1118 surgical specimens and effects of adjuvant gefitinib and erlotinib. J Thorac Oncol 7(12):1815–1822. https://doi.org/10.1097/JTO.0b013e31826bb7b2
Zhang Z, Wang T, Zhang J et al (2014) Prognostic value of epidermal growth factor receptor mutations in resected non-small cell lung cancer: a systematic review with meta-analysis. PLoS ONE 9(8):e106053. https://doi.org/10.1371/journal.pone.0106053
Long C, Li K, Liu Z et al (2023) Real-world analysis of the prognostic value of EGFR mutation detection in plasma ctDNA from patients with advanced non-small cell lung cancer. Cancer Med. https://doi.org/10.1002/cam4.5582
Kuan FC, Kuo LT, Chen MC et al (2015) Overall survival benefits of first-line EGFR tyrosine kinase inhibitors in EGFR-mutated non-small-cell lung cancers: a systematic review and meta-analysis. Br J Cancer 113(10):1519–1528. https://doi.org/10.1038/bjc.2015.356
Hochmair MJ, Morabito A, Hao D et al (2019) Sequential afatinib and osimertinib in patients with EGFR mutation-positive non-small-cell lung cancer: updated analysis of the observational GioTag study. Future Oncol 15(25):2905–2914. https://doi.org/10.2217/fon-2019-0346
Furuyama K, Harada T, Iwama E et al (2013) Sensitivity and kinase activity of epidermal growth factor receptor (EGFR) exon 19 and others to EGFR-tyrosine kinase inhibitors. Cancer Sci 104(5):584–589. https://doi.org/10.1111/cas.12125
Ke EE, Zhou Q, Zhang QY et al (2017) A higher proportion of the EGFR T790M mutation may contribute to the better survival of patients with exon 19 deletions compared with those with L858R. J Thorac Oncol 12(9):1368–1375. https://doi.org/10.1016/j.jtho.2017.05.018
Nosaki K, Satouchi M, Kurata T et al (2016) Re-biopsy status among non-small cell lung cancer patients in Japan: a retrospective study. Lung Cancer 101:1–8. https://doi.org/10.1016/j.lungcan.2016.07.007
Li B, Sun SZ, Yang M et al (2015) The correlation between EGFR mutation status and the risk of brain metastasis in patients with lung adenocarcinoma. J Neurooncol 124(1):79–85. https://doi.org/10.1007/s11060-015-1776-3
Chang WY, Wu YL, Su PL et al (2018) The impact of EGFR mutations on the incidence and survival of stages I to III NSCLC patients with subsequent brain metastasis. PLoS ONE 13(2):e0192161. https://doi.org/10.1371/journal.pone.0192161
Wu YL, Ahn MJ, Garassino MC et al (2018) CNS efficacy of Osimertinib in patients with T790M-Positive Advanced Non-Small-Cell Lung Cancer: Data from a Randomized Phase III Trial (AURA3). J Clin Oncol 36(26):2702–2709. https://doi.org/10.1200/JCO.2018.77.9363
Grommes C, Oxnard GR, Kris MG et al (2011) Pulsatile” high-dose weekly erlotinib for CNS metastases from EGFR mutant non-small cell lung cancer. Neuro Oncol 13(12):1364–1369. https://doi.org/10.1093/neuonc/nor121
Acknowledgements
Authors 3, 4, and 5 are Ph.D. students working at the Gujarat Cancer & Research Institute affiliated with Gujarat University. Bhoomi Tarapara, Hitarth Patel and Hunayna Bhavnagari are Ph.D. students working at the Gujarat Cancer & Research Institute affiliated with Gujarat University.
Funding
No funds, grants, or other support were received for the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. FDS: has conceptualized the study. Material preparation, data collection was performed by BT, HP and HB. Data analysis was done by JJ and AP. The first draft of the manuscript was written by JJ and all authors commented on previous versions of the manuscript. Review and editing were done by FDS. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
As it is a retrospective observational study, no ethical approval is required.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Joshi, J., Pandit, A., Tarapara, B. et al. An association of epidermal growth factor receptor mutation subtypes with prognostic prediction and site-specific recurrence in advanced stage lung cancer patients. Mol Biol Rep 50, 5105–5115 (2023). https://doi.org/10.1007/s11033-023-08432-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08432-2