Abstract
Background
The dissemination of carbapenem resistance via carbapenemases, such as the metallo-β-lactamase NDM, among Enterobacterales poses a public health threat. The aim of this study was to characterize a plasmid carrying the blaNDM-1 gene, which was extracted from a clinical Klebsiella pneumoniae uropathogen from an Egyptian patient suffering from a urinary tract infection.
Methods and results
The recovered plasmid was transformed into competent E. coli DH5α which acquired phenotypic resistance to cefoxitin, ceftazidime, and ampicillin/sulbactam, and intermediate sensitivity to ceftriaxone and imipenem (a carbapenem). Whole plasmid sequencing was performed on the extracted plasmid using the DNBSEQ™ platform. The obtained forward and reverse reads were assembled into contigs using the PRINSEQ and PLACNETw web tools. The obtained contigs were uploaded to PlasmidFinder and ResFinder for in silico plasmid typing and detection of antimicrobial resistance genes, respectively. The final consensus sequence was obtained using the Staden Package software. The plasmid (pNDMKP37, NCBI accession OK623716.1) was typed as an IncX3 plasmid with a size of 46,160 bp and harbored the antibiotic resistance genes blaNDM-1, bleMBL, and aph(3’)-VI. The plasmid also carried mobile genetic elements involved in the dissemination of antimicrobial resistance including insertion sequences IS30, IS630, and IS26.
Conclusions
This is Egypt’s first report of a transmissible plasmid co-harboring blaNDM-1 and aph(3’)-VI genes. Moreover, the respective plasmid is of great medical concern as it has caused the horizontal transmission of multidrug-resistant phenotypes to the transformant. Therefore, new guidelines should be implemented for the rational use of broad-spectrum antibiotics, particularly carbapenems.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Antimicrobial resistance (AMR) has been on the rise globally over the last decade, in addition to a shortage of functional antimicrobials, and a lack of novel ones. Such resistance has a major impact on treatment outcomes, resulting in higher antimicrobial costs, longer hospital stays, greater hospital expenses, and higher fatality rates [1]. In an effort to overcome the increasing rates of resistance to penicillins, cephalosporins, aminoglycosides, and fluoroquinolones, the use of carbapenems has increased in recent years [2, 3]. Regrettably, the extensive usage of carbapenems has led to the development of carbapenem-resistance (CR) particularly, in Gram-negative bacteria (GNB) such as Enterobacterales, Acinetobacter spp., and Pseudomonas spp. This has resulted in a global public health crisis due to the rapid spread of CR and the scarcity of novel antimicrobials [4,5,6].
The World Health Organization (WHO) announced in 2017 a global priority pathogens list for which new antibiotics are urgently needed, among which carbapenem-resistant Enterobacterales (CRE) that are co-resistant to 3rd generation cephalosporins were listed as critical priority pathogens [7]. Infections caused by CRE pathogens have caused substantial morbidity and mortality, and are considered as a rising healthcare threat [8, 9]. Carbapenemase-producing K. pneumoniae represents the fastest-growing threat to antibiotic resistance in terms of morbidity and mortality [10,11,12].
Among the different mechanisms of CR, carbapenemases represent the greatest threat in terms of AMR spread, due to their ability to inactivate most β-lactams, and the fact that they are encoded by genes conferred by mobile genetic elements (MGEs) such as transposons, insertion sequences, or plasmids, which are capable of interspecies and intraspecies horizontal transfer [13, 14]. Plasmids are extrachromosomal, self-replicating DNA units that encode nonessential but usually useful traits for their host. Several copies of one or more plasmids may be present within a bacterial cell [15]. Many plasmids encode genes for resistance to antimicrobial agents and heavy metals, virulence factors, production of toxins, attachment to intestinal mucosa, and for new pathways of degradation [16]. Acquisition of such plasmids enables the host bacterium to adapt to environmental changes, such as exposure to antibiotics, rapidly and effectively [17]. Plasmid replicon typing was established in order to facilitate their identification and study [18].
The New Delhi metallo-β-lactamase (NDM), a carbapenemase that belongs to Ambler class B β-lactamases, was discovered in 2008 when a Swedish patient who had traveled to New Delhi, India, acquired a urinary tract infection caused by a carbapenem-resistant K. pneumoniae. The strain isolated from his urine was a metallo-β-lactamase (MBL) producer but was negative for previously known MBL genes. This led to the discovery of the blaNDM-1 gene [19]. It was later revealed that NDM β-lactamases confer resistance to almost all β-lactam antimicrobials (except aztreonam), including carbapenems which are often used as last-resort treatment options for multidrug-resistant (MDR) and extended-spectrum β-lactamase (ESBL) producers-associated infections [20, 21].
Genes coding for NDM enzymes are highly transmissible as they are often located on plasmids harboring several other antimicrobial resistance determinants, thus, NDM-producing bacteria are often resistant to aminoglycosides and fluoroquinolones, a fact which has posed challenges in the clinical treatment of infections caused by CRE [20, 22, 23].
In this study, we report the sequence of a transmissible IncX3 plasmid carrying a carbapenemase resistance gene (blaNDM-1), bleomycin resistance gene (bleMBL), and aminoglycoside resistance gene (aph(3’)-VI), which was extracted from a carbapenem-resistant K. pneumoniae clinical isolate recovered from the urine of an Egyptian patient suffering from a urinary tract infection.
Materials and methods
Collection of the clinical isolate
Carbapenem-resistant K. pneumoniae isolate (code: 37.AK) was obtained from El-Demerdash Tertiary Care Hospital’s Microbiology laboratory in Cairo, Egypt. According to the hospital’s records, the isolate was recovered from the urine sample of a male patient admitted to the hospital with significant bacteriuria (uropathogens > 105 cfu/ml) in May 2020. The identification of the isolate was carried out using phenotypic and cultural characteristics [24]. Identification of the isolate was further confirmed by using the commercially available MIKROLATEST® ID Kit ENTEROtest 24 N (Erba Lachema, The Czech Republic) [25] following the manufacturer’s instructions. The patient had no history of international travel. Written and oral informed consents were attained from the patient. The study was approved by the Ethics Committee of the Faculty of Pharmacy Ain Shams University (EN-REC-ASU-2019-98) and was in accordance with the Declaration of Helsinki.
Antimicrobial susceptibility testing
The Kirby-Bauer disk diffusion test was done [26] using 17 antimicrobials including meropenem (10 µg), imipenem (10 µg), doripenem (10 µg), ertapenem (10 µg), ampicillin/sulbactam (20 µg), amoxicillin/clavulanic acid (30 µg), ceftriaxone (30 µg), cefoxitin (30 µg), ceftazidime (30 µg), cefepime (30 µg), ciprofloxacin (5 µg), levofloxacin (5 µg), amikacin (30 µg), gentamicin (10 µg), trimethoprim/sulfamethoxazole (25 µg), fosfomycin (200 µg), and nitrofurantoin (300 µg). After incubation, the inhibition zone diameters were measured and susceptibility was interpreted by referring to Clinical and Laboratory Standards Institute (CLSI) guidelines 2020 [27]. The reference strain E. coli ATCC® 25,922 was used as control.
Detection of carbapenemase production
Blue-Carba test was performed as described by Pires et al. for the detection of carbapenemases directly from bacterial cultures [28]. The modified carbapenem inactivation method (mCIM) was also used to detect the production of carbapenemases as recommended by the CLSI 2020 [27]. The reference strain E. coli ATCC® 25,922 was used as control.
Amplification of some plasmid-encoded carbapenemase genes
The plasmid DNA was extracted using the GeneJet Plasmid Miniprep Kit (catalog number: K0502 Thermo Fisher Scientific, Lithuania). The blaNDM, blaKPC, blaOXA-48,blaVIM, and blaIMP carbapenemase genes were amplified using the plasmid extract as a template for PCR and the appropriate primers synthesized by Invitrogen® (Thermo Fisher Scientific, UK), and DreamTaq™ Green PCR Master Mix (Thermo Fisher Scientific, Lithuania). Gel electrophoresis was used to analyze the PCR products as previously reported [29]. Table 1 shows the primers used for PCR amplification of the tested carbapenemase-encoding genes, their annealing temperatures, and the expected product sizes of the tested genes.
Transformation
Chemical transformation was performed according to the protocol described by Sambrook and Russell [29]. E. coli DH5α is a standard susceptible strain free from antimicrobial resistance genes. The preparation of competent E. coli DH5α cells was carried out via the modified Hanahan method [30]. The plasmid extract was used to transform the competent E. coli DH5α. The transformant was cultured on an LB/ampicillin agar plate at a concentration of 100 µg/ml. Untransformed E.coli DH5α was used as a negative control. Blue-Carba test, mCIM test, and antimicrobial susceptibility testing of the transformant were carried out, as well as the determination of the minimum inhibitory concentrations (MICs) of meropenem and imipenem by broth microdilution method following CLSI guidelines [27, 31]. The EDTA-modified carbapenem inactivation method (eCIM) is a phenotypic test used to differentiate MBLs (e.g. NDM, VIM, and IMP carbapenemases) from serine carbapenemases (e.g. OXA-48, and KPC carbapenemases) in Enterobacterales isolates showing positive mCIM test results. This test was carried out and interpreted as mentioned in the CLSI guidelines [27] to determine the type of carbapenemase enzyme produced by the transformant. The plasmid DNA was then extracted from the transformant (TS37.AK) to be used as a template for PCR amplification for confirming the presence of carbapenemase genes on the transformed plasmid using the primers listed in Table 1.
Plasmid sequencing and bioinformatic analysis
The transformant was sub-cultured 2 successive times on LB/ampicillin (100 µg/ml), followed by a third time on LB/meropenem (4 µg/ml), to increase the copy number of the plasmid by positive pressure. The plasmid DNA was then extracted from the transformant, assigned the name pNDMKP37, and was sent for whole plasmid sequencing at BGI TECH SOLUTIONS (HONGKONG) CO., LIMITED (Tai Po, Hong Kong) using DNBSEQ™ platform with PE100. The obtained clean reads were assembled into contigs using the PReprocessing and INformation of SEQuences (PRINSEQ) v0.20.4 (http://prinseq.sourceforge.net/) (accessed on 15 December 2022) and PLACNETw (https://castillo.dicom.unican.es/upload/) (accessed on 15 December 2022) web tools [32, 33]. The obtained contigs were uploaded to PlasmidFinder 2.1 (https://cge.cbs.dtu.dk/services/PlasmidFinder/) (accessed on 15 December 2022) for in silico plasmid typing. ResFinder 4.1 (https://cge.cbs.dtu.dk/services/ResFinder/) (accessed on 15 December 2022) and the Comprehensive Antibiotic Resistance Database (CARD) (https://card.mcmaster.ca/)(accessed on 15 December 2022) were used to detect antimicrobial resistance genes. The consensus sequence was finally assembled using the Staden Package Gap4 software v2.0.0b11 (http://staden.sourceforge.net/)(accessed on 15) [34]. The constructed plasmid was automatically annotated via Bacterial and Viral Bioinformatics Resource Center (BV-BRC) (https://www.bv-brc.org/) (accessed on 17 December 2022) followed by manual inspection for confirmation and correction using ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/)(accessed on 17 December 2022) and BLAST tools (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (accessed on 17 December 2022).
The plasmid sequence was uploaded to the plasmid database PLSDB (https://ccb-microbe.cs.uni-saarland.de/plsdb/) (accessed on 3 December 2022) to search the database for plasmids with nucleotide sequences similar to our plasmid (pNDMKP37). The search strategy used was “Mash dist.” (for long sequences e.g., contigs or long reads), and the parameters were maximum p-value of 0.05 and maximum distance of 0.05. The creation of the circular image and comparison with other reported similar plasmids were performed using the BLAST Ring Image Generator (BRIG) tool v0.95 (https://sourceforge.net/projects/brig/)(accessed on 3 December 2022) [35].
Nucleotide sequence accession number and data availability
The plasmid pNDMKP37 sequence project has been deposited in the GenBank under BioProject PRJNA878540, sample number SAMN30732688, and with the plasmid sequence accession number OK623716.1. The Sequence Read Archive (SRA) data are available from GenBank under the accession number SRR21492341.
Results
Antimicrobial susceptibility, carbaenemase-production, and PCR amplification of carbapenemase-encoding genes
The results of antimicrobial susceptibility, Blue-Carba test, mCIM, and PCR amplification of carbapenemase-encoding genes of the carbapenem-resistant K. pneumoniae clinical isolate (37.AK) were previously published [36]. In terms of antimicrobial susceptibility to carbapenems, the isolate was resistant to meropenem, imipenem, and ertapenem, and was susceptible to doripenem.
Transformation
The transformant (TS37.AK) showed growth on an LB/ampicillin agar plate (100 µg/ml), indicating the successful transformation of a plasmid harboring at least one β-lactam resistance gene. Antimicrobial susceptibility results showed that the transformant (TS37.AK) acquired a new antimicrobial resistance profile compared to the untransformed E. coli DH5α, where this plasmid conferred phenotypic resistance to ceftazidime, cefoxitin, and ampicillin/sulbactam, and intermediate sensitivity to ceftriaxone and imipenem in the transformant. The transformant remained susceptible to ertapenem and doripenem. The MICs of meropenem and imipenem against the transformant were 0.5 µg/ml (susceptible) and 2 µg/ml (intermediate sensitivity), respectively. The results of Blue-Carba and mCIM tests were positive for the transformant indicating carbapenemase-production. The results of eCIM test confirmed that the transformant was an MBL producer, where the carbapenemase inhibition test with EDTA showed a complete inhibition of the carbapenemase activity indicating that the transformant was a sole MBL producer. The phenotypic characteristics of the transformant (TS37.AK), the respective parent K. pneumoniae clinical isolate (37.AK), and the untransformed E. coli DH5α were previously published [36] and are demonstrated in Table 2. After the plasmid DNA was extracted from the transformant, the results of PCR amplification of carbapenemase-encoding genes confirmed that the transformant was carrying blaOXA-48 and blaNDM genes (Figures S1 and S2).
Plasmid sequencing and bioinformatic analysis
The assembled consensus sequences
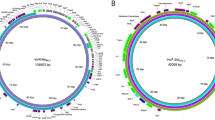
Clean forward and reverse sequence reads were assembled into 6 contigs. The plasmid belonged to the IncX3 plasmid incompatibility group and was found to carry 3 resistance genes, namely blaNDM-1 coding for NDM-1 carbapenemase, bleMBL coding for bleomycin resistance protein BRPMBL, and aph(3’)-VI gene coding for APH(3’)-VI (aminoglycoside 3′- phosphotransferase). The final obtained consensus sequence length was 46,160 bp with 46.28% G + C content. The plasmid contained 58 open reading frames (ORFs) (37 genes of known functions and 21 genes of hypothetical proteins, Fig. 1). The insertion sequence IS30 family transposase (98.98% similarity to ISAba125) was found upstream of blaNDM-1, and bleomycin resistance gene, bleMBL, was located downstream. The genes dsb and trpF were found downstream of bleMBL, all of which are common genetic contexts of blaNDM gene variants [37]. Other MGEs were found in the genetic environment of blaNDM-1, including the insertion sequences IS630 and IS26, which can promote the mobilization of blaNDM-1 between plasmids or chromosomes.
The circular map of the NDM-positive plasmid pNDMKP37 (K. pneumoniae strain 37.AK; AC, OK623716.1) compared to other reported similar plasmids from different Enterobacterales strains, including pNDM-EC36 (E. coli strain EC36; AC, NZ_MG591703.1), pNDM5_020046 (K. pneumoniae subsp. pneumoniae strain SCKP020046; AC, CP028781.2), p3804_NDM (Enterobacter hormaechei strain 3804; AC, CP064660.1), pRIVM_C018652_2 (K. pneumoniae strain RIVM_C018652; AC, CP068835.1), and pNDM5-LDR (K. pneumoniae strain LDR; AC, MK308632.1). The red ring represents the plasmid used as a reference for the alignment (pNDMKP37); the size of the reference is indicated in the center of the panel. The genes coding for APH (3ʹ)-VI, NDM-1, and BRPMBL proteins are marked in orange in the outer ring, and hypothetical proteins are marked in blue. AC, GenBank accession code
Discussion
The most effective antimicrobials for treating infections caused by MDR bacteria are carbapenems. This class of antimicrobials exhibits a wide spectrum of activity against both Gram-negative and Gram-positive bacteria. The overuse of carbapenems in many nations has accelerated the emergence of carbapenem resistance, resulting in a worldwide public health crisis [38]. Increasing numbers of Enterobacterales (especially Klebsiella spp.) and lactose non-fermenters are acquiring and producing carbapenemases. NDMs, which confer resistance to carbapenems and other β-lactam antibiotics, have been increasingly reported across the world since their first report.
In this study we report an IncX3 transmissible plasmid carrying 3 resistance genes: the carbapenemase resistance gene (blaNDM-1) coding for NDM-1 carbapenemase, bleomycin resistance gene (bleMBL) coding for bleomycin resistance protein BRPMBL, and aminoglycoside resistance gene (aph(3’)-VI) coding for aminoglycoside 3′- phosphotransferase APH(3’)-VI. The plasmid was extracted from a carbapenem-resistant K. pneumoniae clinical isolate recovered from the urine of an Egyptian patient suffering from a urinary tract infection. In our previous study involving the same transformant (TS37.AK), the PCR amplification results of the transferable plasmid revealed the presence of genes coding for carbapenemases in the plasmid extract. However, after whole plasmid sequencing in the current study, the sequencing data confirmed that this plasmid carried the blaNDM-1 gene coding for NDM-1 carbapenemase, as well as other resistance determinants namely the bleMBL gene coding for bleomycin resistance protein BRPMBL, and aph(3’)-VI gene coding for APH(3’)-VI (aminoglycoside 3′- phosphotransferase).
Generally, the blaNDM gene variants share two common features pertaining to their genetic environment: the bleomycin resistance gene (bleMBL) is always downstream of blaNDM, and the ISAba125 insertion sequence (either intact or truncated) is always upstream. Further downstream of bleMBL, there is often a set of several genes, including trpF (which encodes a phosphoribosylanthranilate isomerase), and dsb (which encodes a twin-arginine translocation pathway signal sequence domain protein) [37]. This was evident in the sequence of the plasmid in the present study (pNDMKP37), where bleMBL was located downstream of blaNDM-1, and the insertion sequence IS30 family transposase (98.98% similarity to ISAba125) was found upstream. Additionally, the genes trpF and dsb were found downstream of bleMBL. Other MGEs were found in the genetic context of blaNDM-1, including insertion sequences IS630 and IS26, which can promote the mobilization of blaNDM-1 between plasmids or chromosomes.
As for the blaOXA-48 gene, it is generally associated with an upstream IS1999 element, and a downstream composite transposon Tn1999 [39]. Other blaOXA-48-like genes have been found on plasmids in association with other insertion sequences, for example the blaOXA-163 gene was located downstream of an ISEcl4 element, and blaOXA-181, blaOXA-204, and blaOXA-232 genes were associated with ISEcp1 elements [39]. In our study, although blaOXA-48 gene was detected by PCR in the plasmid extract of the transformant (TS37.AK), neither the gene nor its usual genetic context were detected on the sequenced plasmid. To confirm that the blaOXA-48 gene not being in the plasmid was not a bioinformatics processing error, whole genome DNA templates of the transformant were prepared as described by Doyle et al. [40], and the supernatant was used as a template for PCR amplification of blaOXA-48 and blaNDM genes. This was done to detect the possibility that the blaOXA-48 could be carried on a MGE that was not incorporated in the sequenced plasmid and was instead chromosomally incorporated. The PCR results confirmed the presence of the blaNDM gene and the absence of the blaOXA-48 gene from the whole genome of the transformant, although the blaOXA-48 gene was previously detected in the initial plasmid extract of the transformant by PCR and subsequent sanger sequencing of the resulting amplicon [36]. It was thus speculated that the MGE carrying the blaOXA-48 gene might have been lost from our plasmid after repeated subculturing of the transformant, which could be proved by whole genome sequencing of the clinical isolate and the transformant.
The PLSDB plasmid database was searched for plasmids having nucleotide sequences similar to our plasmid (pNDMKP37). The results of searching PLSDB database showed 397 hits, of which 250 (62.97%) plasmids carried blaNDM gene variants as follows: 63 plasmids carried the blaNDM-1, 9 carried the blaNDM-4, 143 carried blaNDM-5, 1 harbored blaNDM-6, 27 carried the blaNDM-7, 1 carried blaNDM-11, 1 harbored blaNDM-13, 1 harbored blaNDM-17, 2 harbored blaNDM-19, 1 carried blaNDM-20, and 1 carried blaNDM-21 variant. It was noticed that similar to our plasmid (pNDMKP37), 246/250 plasmids (98.4%) carrying the blaNDM gene variants also harbored the bleomycin resistance determinant ble. Seventy of the similar plasmids (17.6%) carried the blaOXA-181 resistance determinant, and only 1 of which (plasmid pRIVM_C018652_2, accession no: NZ_CP068835.1) carried both blaNDM-5 (a blaNDM variant), and blaOXA-181 (a blaOXA-48 variant) genes together. This plasmid was extracted from a K. pneumoniae isolate as well. When the nucleotide sequences of both plasmids were compared, it was found that the blaOXA-181 gene in plasmid pRIVM_C018652_2 was carried on a MGE (as shown in Fig. 2) whose sequence was missing from our plasmid (pNDMKP37). It was also noted that the size of pRIVM_C018652_2 (69,764 bp) was larger than that of pNDMKP37 (46,160 bp).
The circular map of the NDM-positive plasmid pNDMKP37 (K. pneumoniae strain 37.AK; AC, OK623716.1) compared to the OXA-181 and NDM-positive plasmid (K. pneumoniae strain RIVM_C018652; AC, NZ_CP068835.1). The blue ring represents the plasmid used as a reference for the alignment (pRIVM_C018652_2); the size of the reference is indicated in the center of the panel. The genes on plasmid pRIVM_C018652_2 coding for NDM-5, BRPMBL, and OXA-181 proteins are marked in orange in the outer ring. Transposases are marked in black. AC, GenBank accession code
It was noticed that the parent K. pneumoniae clinical isolate (37.AK) was resistant to aminoglycosides (gentamicin and amikacin) and quinolones (ciprofloxacin and levofloxacin), however, such resistance was not conferred to the transformant by the transformed plasmid. This is probably because resistance mechanisms to such antimicrobial agents was chromosomally-mediated in the parent clinical isolate.
Conclusions
In this study, a transmissible plasmid co-harboring blaNDM-1 and aph(3’)-VI genes was detected in a K. pneumoniae clinical isolate in Egypt. The plasmid (pNDMKP37) was typed as an IncX3 plasmid with a size of 46,160 bp and harbored antibiotic resistance genes against carbapenems (blaNDM-1), bleomycin (bleMBL), and aminoglycosides (aph(3’)-VI). Upon transformation of the pNDMKP37 in E. coli DH5α, it conferred phenotypic resistance against ceftazidime, cefoxitin, and ampicillin/sulbactam, and intermediate sensitivity to ceftriaxone and imipenem. Moreover, the respective plasmid poses a great medical concern as it was responsible for the horizontal transmission of multidrug-resistant phenotypes to the transformant, particularly phenotypic resistance to carbapenems. New guidelines should be implemented for the rational use of broad-spectrum antibiotics, particularly carbapenems.
Data Availability
All the data supporting the findings are included in the manuscript. The plasmid pNDMKP37 sequence project has been deposited in the GenBank under BioProject PRJNA878540, sample number SAMN30732688, and with the plasmid sequence accession number OK623716.1. The Sequence Read Archive (SRA) data are available from GenBank under the accession number SRR21492341.
Acknowledgments:
Abbreviations
- AMR:
-
Antimicrobial resistance
- CLSI:
-
Clinical and Laboratory Standards Institute
- CR:
-
Carbapenem resistance
- CRE:
-
Carbapenem-resistant
- Enterobacterales, eCIM:
-
EDTA-modified carbapenem inactivation method
- ESBL:
-
Extended-spectrum -lactamase
- GNB:
-
Gram-negative bacteria
- MBL:
-
Metallo--lactamase
- mCIM:
-
Modified carbapenem inactivation method
- MDR:
-
Multidrug-resistant
- MGE:
-
Mobile genetic element
- NDM:
-
New Delhi metallo--lactamase
- ORF:
-
Open reading frame
- PCR:
-
Polymerase chain reaction
- WHO:
-
World Health Organization
References
Sleiman A, Fayad AGA, Banna H, Matar GM (2021) Prevalence and molecular epidemiology of carbapenem-resistant Gram-negative bacilli and their resistance determinants in the Eastern Mediterranean Region over the last decade. J Glob Antimicrob Resist 25:209–221. https://doi.org/10.1016/j.jgar.2021.02.033
Schwaber MJ, Navon-Venezia S, Schwartz D, Carmeli Y (2005) High levels of Antimicrobial Coresistance among extended-Spectrum-β-Lactamase-producing Enterobacteriaceae. Antimicrob Agents Chemother 49:2137–2139. https://doi.org/10.1128/AAC.49.5.2137-2139.2005
Zhanel G, Hisanaga T, Laing N et al (2006) Antibiotic resistance in Escherichia coli outpatient urinary isolates: final results from the north american urinary tract infection Collaborative Alliance (NAUTICA). Int J Antimicrob Agents 27:468–475. https://doi.org/10.1016/j.ijantimicag.2006.02.009
Diene SM, Rolain JM (2014) Carbapenemase genes and genetic platforms in Gram-negative bacilli: Enterobacteriaceae, Pseudomonas and Acinetobacter species. Clin Microbiol Infect 20:831–838. https://doi.org/10.1111/1469-0691.12655
Harris PNA, Tambyah PA, Paterson DL (2015) β-lactam and β-lactamase inhibitor combinations in the treatment of extended-spectrum β-lactamase producing Enterobacteriaceae: time for a reappraisal in the era of few antibiotic options? Lancet Infect Dis 15:475–485. https://doi.org/10.1016/S1473-3099(14)70950-8
Codjoe F, Donkor E (2017) Carbapenem Resistance: a review. Med Sci 6:1. https://doi.org/10.3390/medsci6010001
WHO | Prioritization of pathogens to guide discovery, research and development of new antibiotics for drug resistant bacterial infections, including tuberculosis.WHO
Sheu C-C, Chang Y-T, Lin S-Y et al (2019) Infections caused by Carbapenem-Resistant Enterobacteriaceae: an update on Therapeutic Options. Front Microbiol 10. https://doi.org/10.3389/fmicb.2019.00080
Logan LK, Weinstein RA (2017) The epidemiology of Carbapenem-Resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis 215:S28–S36. https://doi.org/10.1093/infdis/jiw282
Brink AJ (2019) Epidemiology of carbapenem-resistant Gram-negative infections globally. Curr Opin Infect Dis 32:609–616. https://doi.org/10.1097/QCO.0000000000000608
Lee C-R, Lee JH, Park KS et al (2016) Global dissemination of carbapenemase-producing Klebsiella pneumoniae: Epidemiology, genetic context, Treatment Options, and detection methods. Front Microbiol 7:1–30. https://doi.org/10.3389/fmicb.2016.00895
Karampatakis T, Tsergouli K, Behzadi P (2023) Carbapenem-Resistant Klebsiella pneumoniae: virulence factors, molecular epidemiology and latest updates in Treatment Options. Antibiotics 12:234. https://doi.org/10.3390/antibiotics12020234
Partridge SR, Kwong SM, Firth N, Jensen SO (2018) Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin Microbiol Rev 31:1–61. https://doi.org/10.1128/CMR.00088-17
Kamel NA, Tohamy ST, Yahia IS, Aboshanab KM (2022) Insights on the performance of phenotypic tests versus genotypic tests for the detection of carbapenemase-producing Gram-negative bacilli in resource-limited settings. BMC Microbiol 22:248. https://doi.org/10.1186/s12866-022-02660-5
Caugant DA (2009) Molecular Epidemiology of Microorganisms. Humana Press, Totowa, NJ
Streips UN, Yasbin RE (2002) Modern Microbial Genetics, 2nd edn. Wiley-Liss, New York, NY
Smets BF, Barkay T (2005) Horizontal gene transfer: perspectives at a crossroads of scientific disciplines. Nat Rev Microbiol 3:675–678. https://doi.org/10.1038/nrmicro1253
Erdem F, Oncul O, Aktas Z (2019) Characterization of resistance genes and polymerase chain reaction-based replicon typing in Carbapenem-Resistant Klebsiella pneumoniae. Microb Drug Resist 25:551–557. https://doi.org/10.1089/mdr.2018.0231
Yong D, Toleman MA, Giske CG et al (2009) Characterization of a new metallo-β-lactamase gene, blaNDM–1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53:5046–5054. https://doi.org/10.1128/AAC.00774-09
Findlay J, Poirel L, Kessler J et al (2021) New Delhi Metallo-β-Lactamase–producing Enterobacterales Bacteria, Switzerland, 2019–2020. Emerg Infect Dis 27:2628–2637. https://doi.org/10.3201/eid2710.211265
Meletis G (2016) Carbapenem resistance: overview of the problem and future perspectives. Ther Adv Infect Dis 3:15–21. https://doi.org/10.1177/2049936115621709
Chen Q, Zhou J, Wu S et al (2020) Characterization of the IncX3 plasmid producing blaNDM–7 from Klebsiella pneumoniae ST34. Front Microbiol 11:1–7. https://doi.org/10.3389/fmicb.2020.01885
Jain A, Hopkins KL, Turton J et al (2014) NDM carbapenemases in the United Kingdom: an analysis of the first 250 cases. J Antimicrob Chemother 69:1777–1784. https://doi.org/10.1093/jac/dku084
Bergey DH, Holt JG (1994) Bergey’s manual of determinative bacteriology, 9th edn. Williams & Wilkins, Baltimore
Uvarova YE, Bryanskaya AV, Rozanov AS et al (2020) An integrated method for taxonomic identif ication of microorganisms. Vavilov J Genet Breed 24:376–382. https://doi.org/10.18699/VJ20.630
CLSI (2018) Performance standards for antimicrobial disk susceptibility tests. 13th ed. CLSI Standard M02, 13th ed. Clinical and Laboratory Standards Institute, Wayne, PA
CLSI (2020) Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. CLSI supplement M100, 30th ed. Clinical and Laboratory Standards Institute, Wayne, PA
Pires J, Novais A, Peixe L (2013) Blue-Carba, an easy biochemical test for detection of Diverse Carbapenemase Producers directly from bacterial cultures. J Clin Microbiol 51:4281–4283. https://doi.org/10.1128/JCM.01634-13
Sambrook J, Russell DW (2001) Molecular cloning: a Laboratory Manual, 3rd edn. Cold Spring Harbor Laboratory Press, New York
Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. https://doi.org/10.1016/S0022-2836(83)80284-8
CLSI (2018) Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 11th ed. CLSI standard M07, 11th ed. Clinical and Laboratory Standards Institute, Wayne, PA
Vielva L, de Toro M, Lanza VF, de la Cruz F (2017) PLACNETw: a web-based tool for plasmid reconstruction from bacterial genomes. Bioinformatics 33:3796–3798. https://doi.org/10.1093/bioinformatics/btx462
de Toro M, Lanza VF, Vielva L et al (2020) Plasmid Reconstruction from Next-Gen Data: a detailed protocol for the Use of PLACNETw for the Reconstruction of plasmids from WGS datasets. In: de la Cruz F (ed) Horizontal gene transfer. Springer US, New York, NY, pp 323–339
Staden R (1996) The staden sequence analysis package. Mol Biotechnol 5:233–241. https://doi.org/10.1007/BF02900361
Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA (2011) BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. https://doi.org/10.1186/1471-2164-12-402
Elshamy AA, Saleh SE, Alshahrani MY et al (2021) OXA-48 carbapenemase-encoding transferable plasmids of Klebsiella pneumoniae recovered from egyptian patients suffering from complicated urinary tract infections. Biology (Basel) 10:889. https://doi.org/10.3390/biology10090889
Wu W, Feng Y, Tang G et al (2019) NDM Metallo-β-Lactamases and their bacterial producers in Health Care settings. Clin Microbiol Rev 32:e00115–e00118. https://doi.org/10.1128/CMR.00115-18
Aurilio C, Sansone P, Barbarisi M et al (2022) Mechanisms of action of Carbapenem Resistance. Antibiotics 11:421. https://doi.org/10.3390/antibiotics11030421
Evans BA, Amyes SGB (2014) OXA β-Lactamases. Clin Microbiol Rev 27:241–263. https://doi.org/10.1128/CMR.00117-13
Doyle D, Peirano G, Lascols C et al (2012) Laboratory Detection of Enterobacteriaceae that produce carbapenemases. J Clin Microbiol 50:3877–3880. https://doi.org/10.1128/JCM.02117-12
Nordmann P, Poirel L, Carrer A et al (2011) How to detect NDM-1 Producers. J Clin Microbiol 49:718–721. https://doi.org/10.1128/JCM.01773-10
Nordmann P, Naas T, Poirel L (2011) Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 17:1791–1798. https://doi.org/10.3201/eid1710.110655
Poirel L, Naas T, Nicolas D et al (2000) Characterization of VIM-2, a Carbapenem-Hydrolyzing Metallo-β-Lactamase and its plasmid- and Integron-Borne Gene from a Pseudomonas aeruginosa Clinical isolate in France. Antimicrob Agents Chemother 44:891–897. https://doi.org/10.1128/AAC.44.4.891-897.2000
Woodford N, Tierno PM, Young K et al (2004) Outbreak of Klebsiella pneumoniae producing a New Carbapenem-Hydrolyzing Class A β-Lactamase, KPC-3, in a New York Medical Center. Antimicrob Agents Chemother 48:4793–4799. https://doi.org/10.1128/AAC.48.12.4793-4799.2004
Acknowledgements
The authors would like to acknowledge the Microbiology and Immunology Department, Faculty of Pharmacy, Ain Shams University for providing the laboratory facilities for this study. The authors are also grateful to the staff at the microbiology laboratories of El-Demerdash Hospital, Cairo, Egypt, for providing the clinical specimen and data records.
The authors would like to acknowledge the Microbiology and Immunology Department, Faculty of Pharmacy, Ain Shams University for providing the laboratory facilities for this study. The authors are also grateful to the staff at the microbiology laboratories of El-Demerdash Hospital, Cairo, Egypt, for providing the clinical specimen and data records.
Funding
This research did not receive any grant from funding agencies in the public, commercial, or non-profit sectors.
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Author information
Authors and Affiliations
Contributions
Conceptualization, A.A.E., S.E.S., K.M.A., M.M.A. and N.A.H.; methodology, A.A.E., S.E.S., M.M.A. and K.M.A.; writing—original draft preparation, A.A.E. and S.E.S.; writing—review and editing, K.M.A., M.M.A. and N.A.H.; supervision, K.M.A., M.M.A. and N.A.H. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethics approval
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Faculty of Pharmacy Ain Shams University Ethics Committee (ENREC-ASU-2019-98).
Consent to participate
Informed consent was obtained from the patient after explaining the purpose of the study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Elshamy, A.A., Saleh, S.E., Aboshanab, K.M. et al. Transferable IncX3 plasmid harboring blaNDM-1, bleMBL, and aph(3’)-VI genes from Klebsiella pneumoniae conferring phenotypic carbapenem resistance in E. coli. Mol Biol Rep 50, 4945–4953 (2023). https://doi.org/10.1007/s11033-023-08401-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08401-9