Abstract
Background
The narrow genetic diversity of chickpea is a serious impediment to modern cultivar creation. Seed storage proteins (SSPs) are stable and have minimal or no degradation when subjected to isolation and SDS-PAGE.
Methods and results
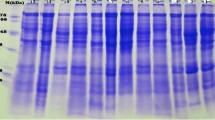
We have characterized SSPs of 436 chickpea genotypes, belonging to nine annual Cicer species, originated from 47 countries by SDS-PAGE and determined the extent of genetic diversity in chickpea through clustering. Based on scoring, a total of 44 bands (10 to 170 kDa) were identified, which were all polymorphic. The least appeared protein bands were 11, 160 and 170 kDa where band of 11 and 160 kDa was present exclusively in wild type. Five bands were present in < 10% of genotypes. Bands appeared in 200–300 genotypes were suggested less polymorphic, on contrary bands present in 10–150 genotypes were suggested more polymorphic. Polymorphism of protein bands in context to their potential functions reported in literature were explored and suggested that the glubulins were most and glutelins were least abundant, whereas albumins with their known role in stress tolerance can be used as marker in chickpea breeding. Cluster analysis produced 14 clusters, interestingly three clusters contained only Pakistani genotypes and thus Pakistani genotypes appeared as a separate entity from the rest of the genotypes.
Conclusion
Our results indicate that SDS-PAGE of SSPs is a powerful technique in determining the genetic diversity plus it is easily adaptable, due to its cost effectiveness in comparison to other genomics tools.





Similar content being viewed by others
Data availability
All data generated or analysed during this study are being included in the manuscript and its supplementary information files.
References
Maiti R, Ebeling WP (2001) Advances in chickpea science. Science Publishers, New York
Siddique K, Bultynck L (2004) Chicpea/Agronomy. In: Wrigley C et al (eds) Encyclopedia of grain science. Marcel Dekker, New York, pp 287–294
Nwokolo E, Smartt JJ (1996) Food and feed from legumes and oilseeds. Chapman & Hall, London
Kaushik I, Singh R, Bhisnoi JP (2017) Effect of barley malt, chickpea and peanut on quality of barley based beverage. J Appl Nat Sci 9:1182–1186
Roy F, Boye J, Simpson B (2010) Bioactive proteins and peptides in pulse crops: pea, chickpea and lentil. Food Res Int 43:432–442
Kaur R, Prasad K (2021) Technological, processing and nutritional aspects of chickpea (Cicer arietinum)-A review. Trends Food Sci Technol 109:448–463
Jukanti AK, Gaur PM, Gowda C, Chibbar RN (2012) Nutritional quality and health benefits of chickpea (Cicer arietinum L.): a review. Br J Nutr 108:S11–S26
Heidarvand L, Maali-Amiri R (2013) Physio-biochemical and proteome analysis of chickpea in early phases of cold stress. J Plant Physiol 170:459–469
Joshi PK, Rao PP (2017) Global pulses scenario: status and outlook. Ann N Y Acad Sci 1392(1):6–17
Gaur PM, Samineni S, Thudi M, Tripathi S, Sajja SB, Jayalakshmi V, Mannur DM, Vijayakumar AG, Ganga Rao NV, Ojiewo C (2019) Integrated breeding approaches for improving drought and heat adaptation in chickpea (Cicer arietinum L.). Plant Breed 138:389–400
Rachwa-Rosiak D, Nebesny E, Budryn G (2015) Chickpeas—Composition, nutritional value, health benefits, application to bread and snacks: a review. Crit Rev Food Sci Nutr 55:1137–1145
Van der Maesen L (1987) Origin, history and taxonomy of chickpea. In: Saxena MC, Singh KB (eds) The chickpea. CAB Int Publ, UK, pp 11–34
Arif MAR, Waheed MQ, Lohwasser U, Shokat S, Alqudah AM, Volkmar C, Börner A (2022) Genetic insight into the insect resistance in bread wheat exploiting the untapped natural diversity. Front Genet 13:828905
Arif A, Parveen N, Waheed MQ, Atif RM, Waqar I, Shah TM (2021) A comparative study for assessing the drought-tolerance of chickpea under varying natural growth environments. Front Plant Sci 11:607869
Li H, Rodda M, Gnanasambandam A, Aftab M, Redden R, Hobson K, Rosewarne G, Materne M, Kaur S, Slater AT (2015) Breeding for biotic stress resistance in chickpea: progress and prospects. Euphytica 204:257–288
Lopes M, Dreisigacker S, Peña R, Sukumaran S, Reynolds MP (2015) Genetic characterization of the wheat association mapping initiative (WAMI) panel for dissection of complex traits in spring wheat. Theor Appl Genet 128:453–464
Kumar P, Yadava RK, Kumar S, Kumar P (2016) Molecular diversity analysis in wheat genotypes using SSR markers. Electron J Plant Breed 7(2):464–468
Ayala FJ, Kiger JA (1984) Modern genetics. The Benjamins/Cummings Publishing Company, Menlo Park, pp 123–134
Körber N, Bus A, Li J, Parkin IA, Wittkop B, Snowdon RJ, Stich B (2016) Agronomic and seed quality traits dissected by genome-wide association mapping in Brassica napus. Front Plant Sci 7:386
Osawaru ME, Ogwu MC, Aiwansoba RO (2015) Hierarchical approaches to the analysis of genetic diversity in plants: a systematic overview. Univ Mauritius Res J 21
Prasad DR, Minocha T, Maurya B, Yousuf PY (2022) Advancement in molecular tools of plant population genetics. Plant ecogenomics. Apple Academic Press, New Jersey, pp 19–45
Rasheed A, Xia X, Yan Y, Appels R, Mahmood T, He Z (2014) Wheat seed storage proteins: advances in molecular genetics, diversity and breeding applications. J Cereal Sci 60(1):11–24
Hassan R, Waheed M, Shokat S, Arif MAR, Tariq R, Arif M, Arif A (2020) Estimation of genomic diversity using sequence related amplified polymorphism (SRAP) markers in a mini core collection of wheat germplasm from Pakistan. Cereal Res Commun 48:33–44
Sayed MR, Alshallash KS, Safhi FA, Alatawi A, Al-shamrani SM, Dessoky ES, Althobaiti AT, Althaqafi MM, Gharib HS, Shafie WW, Awad-Allah MM (2022) Genetic diversity, analysis of some agro-morphological and quality traits and utilization of plant resources of alfalfa. Genes 13(9):1521
Hameed A, Shah TM, Atta BM, Iqbal N, Haq MA, Ali H (2009) Comparative seed storage protein profiling of Kabuli chickpea genotypes. Pak J Bot 41:703–710
Sharma M, Chaudhary M, Raina SN, Sahoo D, Bhavesh NS, Thakur RK, Rajpal VR, Raturi D, Singh A (2023) Comparative analysis of the efficiency of seed protein profiles in assessing genetic variation and population structure among indigenous Manipur black rice cultivars. Mol Biol Rep. https://doi.org/10.1007/s11033-022-08228-w
Tanksley S, Jones R (1981) Application of alcohol dehydrogenase allozymes in testing the genetic purity of F1 hybrids of tomato. Hort Sci 16:179–181
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. https://doi.org/10.1038/227680a0
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Annal Biochem 72:248–254
Kodinariya TM, Makwana PR (2013) Review on determining number of Cluster in K-Means clustering. Int J Adv Res Comput Sci Manage Stud 1(6):90–95
Saputra DM, Saputra D, Oswari LD (2020) Effect of distance metrics in determining k-value in k-means clustering using elbow and silhouette method. SICONIAN.2019. Atlantis Press, Paris, pp 341–346
El-Mandouh AM, Abd-Elmegid LA, Mahmoud HA, Haggag MH (2019) Optimized K-means clustering model based on gap statistic. Int J Adv Comput Sci Appl. https://doi.org/10.14569/IJACSA.2019.0100124
Pal T, Ghosh S, Mondal A, De KK (2016) Evaluation of genetic diversity in some promising varieties of lentil using karyological characters and protein profiling. JGEB 14(1):39–48
Alghamdi SS, Khan MA, Migdadi HM, El-Harty EH, Afzal M, Farooq M (2019) Biochemical and molecular characterization of cowpea landraces using seed storage proteins and SRAP marker patterns. Saudi J Biol Sci 26:74–82
Hameed A, Saddiqa A, Nadeem S, Iqbal N, Atta BM, Shah TM (2012) Genotypic variability and mutant identification in Cicer arietinum L. by seed storage protein profiling. Pak J Bot 44:1303–1310
Wakasa Y, Takaiwa F (2013) The use of rice seeds to produce human pharmaceuticals for oral therapy. Biotechnol J 8:1133–1143
Salunkhe DK, Kadam SS (1989) CRC handbook of world food legumes: nutritional chemistry, processing technology, and utilization. CRC Press, Boca Raton
Vioque J, Clemente A, Sánchez-Vioque R, Pedroche J, Bautista J, Millán F (1998) Comparative study of chickpea and pea PA2 albumins. J Agric Food Chem 46(9):3609–3613
Gueguen J (1991) Pea and fababean proteins. In: Hudson BJF (ed) Development in food proteins. Elsevier, London, pp 35–78
Agrawal L, Narula K, Basu S, Shekhar S, Ghosh S, Datta A, Chakraborty N, Chakraborty S (2013) Comparative proteomics reveals a role for seed storage protein AmA1 in cellular growth, development, and nutrient accumulation. J Proteome Res 12(11):4904–4930
Grasso N, Lynch NL, Arendt EK, O’Mahony JA (2022) Chickpea protein ingredients: a review of composition, functionality, and applications. CRFSFS 21(1):435–452
Gravel A, Doyen A (2023) Pulse globulins 11S and 7S: origins, purification methods, and techno-functional properties. J Agric Food Chem. https://doi.org/10.1021/acs.jafc.2c07507
Plietz B, Damaschum G, Schwenke KD (1980) Quaternary structure of lIS globulin from different plant seeds. Stud Biophys 79:145–146
Singh U, Jambunatha R (1982) Distribution of seed protein fractions and amino acids in different anatomical parts of chickpea (Cicer arietinum L.) and pigeon pea (Cajanus cajan L.). Plant Foods for Hum Nutr 31(10):347–354
Chang YW, Alli I, Molina AT, Konishi Y, Boye JI (2012) Isolation and characterization of chickpea (Cicer arietinum L.) seed protein fractions. Food Bioprocess Technol 5(2):pp618–625
Arif MAR, Bux H, Kazi AG, Rasheed A, Napar AA, Riaz A, Mujeeb-Kazi A (2012) Stripe rust analysis of D-genome synthetic wheats (2 n = 6 x = 42, AABBDD) and their molecular diversity. Arch Phytopathol Pfl 45:1479–1487
Rubio LA, Grant G, Cavalle C, Martinez-Aragon A, Pusztai A (1994) High in vivo (rat) digestibility of faba bean (Vicia faba), lupin (Lupinus angustifolius) and soya bean (Glycine max) soluble globulins. J Sci Food Agric 66:289–292
Pena JM, Lozano JA, Larranaga P (1999) An empirical comparison of four initialization methods for the k-means algorithm. Pattern Recognit Lett 20:1027–1040
Nisar M, Ghafoor A, Khan MR, Ahmad H, Qureshi AS, Ali H (2007) Genetic diversity and geographic relationship among local and exotic chickpea germplasm. Pak J Bot 39:1575–1581
Arif MAR, Nagel M, Neumann K, Kobiljski B, Lohwasser U, Börner A (2012) Genetic studies of seed longevity in wheat using segregation and association mapping approaches. Euphytica 186:1–13
Acknowledgements
The authors are thankful to the Australian Grain Genebank, Victoria, Australia and chickpea group at NIAB, Pakistan for providing the seeds of exotic and Pakistani chickpea cultivars respectively.
Funding
No external funding was received for this work.
Author information
Authors and Affiliations
Contributions
AA conceived the idea; UK and MQW performed the original experiments and carried out the gel visualization and band scoring; MARA, AA, UK, MQW and NP performed the analysis; UK, MQW, AA & MARA wrote the manuscript; NP and AA reviewed the manuscript by providing inputs to improve the manuscript. All authors read and approve the final manuscript.
Corresponding author
Ethics declarations
Competing interest
The authors have no relevant financial and non-financial interest to disclose.
Ethical approval
This article does not contain any studies with animals or humans performed by any of the authors.
Additional information
Uswah Khalid and Muhammad Qandeel Waheed shared first authors.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11033_2023_8358_MOESM5_ESM.png
Supplementary material 5 (PNG 67.7 kb) Sub clusters in cluster 1 where different shaded regions indicate different sub groups. For details, see Table S2
11033_2023_8358_MOESM6_ESM.png
Supplementary material 6 (PNG 30.3 kb) Sub clusters in cluster 2 where different colors indicate different sub groups. For details, see Table S3
11033_2023_8358_MOESM7_ESM.png
Supplementary material 7 (PNG 65.4 kb) Sub clusters in cluster 3 where different colors indicate different sub groups. For details, see Table S4
11033_2023_8358_MOESM8_ESM.png
Supplementary material 8 (PNG 28.0 kb) Sub clusters in cluster 4 where different colors indicate different sub groups. For details, see Table S5
11033_2023_8358_MOESM9_ESM.png
Supplementary material 9 (PNG 46.8 kb) Sub clusters in cluster 5 where different colors indicate different sub groups. For details, see Table S6
11033_2023_8358_MOESM10_ESM.png
Supplementary material 10 (PNG 45.6 kb) Sub clusters in cluster 6 where different colors indicate different sub groups. For details, see Table S7
11033_2023_8358_MOESM11_ESM.png
Supplementary material 11 (PNG 29.1 kb) Sub clusters in cluster 7 where different colors indicate different sub groups. For details, see Table S8
11033_2023_8358_MOESM12_ESM.png
Supplementary material 12 (PNG 30.7 kb) Sub clusters in cluster 8 where different colors indicate different sub groups. For details, see Table S9
11033_2023_8358_MOESM13_ESM.png
Supplementary material 13 (PNG 48.0 kb) Sub clusters in cluster 9 where different colors indicate different sub groups. For details, see Table S10
11033_2023_8358_MOESM14_ESM.png
Supplementary material 14 (PNG 54.0 kb) Sub clusters in cluster 10 where different colors indicate different sub groups. For details, see Table S11
11033_2023_8358_MOESM15_ESM.png
Supplementary material 15 (PNG 49.7 kb) Sub clusters in cluster 11 where different colors indicate different sub groups. For details, see Table S12
11033_2023_8358_MOESM16_ESM.png
Supplementary material 16 (PNG 58.4 kb) Sub clusters in cluster 12 where different colors indicate different sub groups. For details, see Table S13
11033_2023_8358_MOESM17_ESM.png
Supplementary material 17 (PNG 43.8 kb) Sub clusters in cluster 13 where different colors indicate different sub groups. For details, see Table S14
11033_2023_8358_MOESM18_ESM.png
Supplementary material 18 (PNG 44.1 kb) Sub clusters in cluster 14 where different colors indicate different sub groups. For details, see Table S15
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Khalid, U., Waheed, M.Q., Parveen, N. et al. Estimation of genetic diversity using seed storage protein (SSP) profiling in wild and cultivated species of Cicer L. Mol Biol Rep 50, 4175–4185 (2023). https://doi.org/10.1007/s11033-023-08358-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08358-9