Abstract
Background
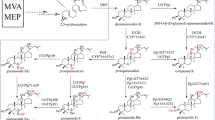
Paris polyphylla var. yunnanensis is an important medicinal plant, and the main active ingredient of the plant is polyphyllin, which is a steroid saponin with pharmacological activities. The central enzyme genes participating in the biosynthesis of polyphyllin are increasingly being uncovered; however, UGTs are rarely illustrated.
Methods and results
In this study, we cloned a new sterol glycosyltransferase from Paris polyphylla var. yunnanensis and identified its catalytic function in vitro. PpUGT6 showed the ability to catalyse the C-3 glycosylation of pennogenin sapogenin of polyphyllin, and PpUGT6 showed catalytic promiscuity towards steroids at the C-17 position of testosterone and methyltestosterone and the triterpene at the C-3 position of glycyrrhetinic acid. Homology modelling of the PpUGT6 protein and virtual molecular docking of PpUGT6 with sugar acceptors and donors were performed, and we predicted the key residues interacting with ligands.
Conclusions
Here, PpUGT6, a novel sterol glycosyltransferase related to the biosynthesis of polyphyllin from P. polyphylla, was characterized. PpUGT6 catalysed C-3 glycosylation to pennogenin sapogenin of polyphyllin, which is the first glycosylation step of the biosynthetic pathway of polyphyllins. Interestingly, PpUGT6 demonstrated glycodiversification to testosterone and methyltestosterone at C-17 and triterpene of glycyrrhetinic acid at the C-3 position. The virtual molecular docking of PpUGT6 protein with ligands predicted the key residues interacting with them. This work characterized a novel SGT glycosylating pennogenin sapogenin at C-3 of polyphyllin from P. polyphylla and provided a reference for further elucidation of the phytosterol glycosyltransferases in catalytic promiscuity and key residues interacting with substrates.





Similar content being viewed by others
References
Tang MJ, Zhao J, Li XH, Yu SS (2004) Advances in studies on chemical constituents and pharmacological activities from plants of Symplocaceae. Zhongguo Zhong Yao Za Zhi 29(5):390–394
Negi JS, Bisht VK, Bhandari AK, Bhatt VP, Singh P, Singh N (2014) Paris polyphylla: chemical and biological prospectives. Anticancer Agents Med Chem 14(6):833–839. https://doi.org/10.2174/1871520614666140611101040
Shuli M, Wenyuan G, Yanjun Z, Chaoyi M, Liu Y, Yiwen L (2011) Paridis saponins inhibiting carcinoma growth and metastasis in vitro and in vivo. Arch Pharm Res 34(1):43–50. https://doi.org/10.1007/s12272-011-0105-4
Qin XJ, Sun DJ, Ni W, Chen CX, Hua Y, He L, Liu HY (2012) Steroidal saponins with antimicrobial activity from stems and leaves of Paris polyphylla var. yunnanensis. Steroids 77(12):1242–1248. https://doi.org/10.1016/j.steroids.2012.07.007
Patel K, Gadewar M, Tahilyani V, Patel DK (2013) A review on pharmacological and analytical aspects of diosmetin: a concise report. Chin J Integr Med 19(10):792–800. https://doi.org/10.1007/s11655-013-1595-3
Guo SY, Yin Y, Lei T, Shi YH, Gao W, Zhang XN, Li J (2021) A cycloartenol synthase from the steroidal saponin biosynthesis pathway of Paris polyphylla. J Asian Nat Prod Res 23(4):353–362. https://doi.org/10.1080/10286020.2020.1730331
Guan HY, Su P, Zhao YJ, Zhang XN, Dai ZB, Guo J, Tong YR, Liu YJ, Hu TY, Yin Y, Gao LH, Gao W, Huang LQ (2018) Cloning and functional analysis of two sterol-C24-methyltransferase 1 (SMT1) genes from Paris polyphylla. J Asian Nat Prod Res 20(7):595–604. https://doi.org/10.1080/10286020.2016.1271791
Yin Y, Gao L, Zhang X, Gao W (2018) A cytochrome P450 monooxygenase responsible for the C-22 hydroxylation step in the Paris polyphylla steroidal saponin biosynthesis pathway. Phytochemistry 156:116–123. https://doi.org/10.1016/j.phytochem.2018.09.005
Christ B, Xu C, Xu M, Li FS, Wada N, Mitchell AJ, Han XL, Wen ML, Fujita M, Weng JK (2019) Repeated evolution of cytochrome P450-mediated spiroketal steroid biosynthesis in plants. Nat Commun 10(1):3206. https://doi.org/10.1038/s41467-019-11286-7
Kurze E, Wust M, Liao J, McGraphery K, Hoffmann T, Song C, Schwab W (2022) Structure-function relationship of terpenoid glycosyltransferases from plants. Nat Prod Rep 39(2):389–409. https://doi.org/10.1039/d1np00038a
Osmani SA, Bak S, Møller BL (2009) Substrate specificity of plant UDP-dependent glycosyltransferases predicted from crystal structures and homology modeling. Phytochemistry 70(3):325–347. https://doi.org/10.1016/j.phytochem.2008.12.009
Schwab W, Fischer TC, Giri A, Wüst M (2015) Potential applications of glucosyltransferases in terpene glucoside production: impacts on the use of aroma and fragrance. Appl Microbiol Biotechnol 99(1):165–174. https://doi.org/10.1007/s00253-014-6229-y
Ullmann P, Ury A, Rimmele D, Benveniste P, Bouvier-Nave P (1993) UDP-glucose sterol beta-D-glucosyltransferase, a plasma membrane-bound enzyme of plants: enzymatic properties and lipid dependence. Biochimie 75(8):713–723. https://doi.org/10.1016/0300-9084(93)90102-x
Webb MS, Irving TC, Steponkus PL (1995) Effects of plant sterols on the hydration and phase behavior of DOPE/DOPC mixtures. Biochim Biophys Acta 1239(2):226–238. https://doi.org/10.1016/0005-2736(95)00147-u
Chaturvedi P, Misra P, Tuli R (2011) Sterol glycosyltransferases–the enzymes that modify sterols. Appl Biochem Biotechnol 165(1):47–68. https://doi.org/10.1007/s12010-011-9232-0
Sharma LK, Madina BR, Chaturvedi P, Sangwan RS, Tuli R (2007) Molecular cloning and characterization of one member of 3beta-hydroxy sterol glucosyltransferase gene family in Withania somnifera. Arch Biochem Biophys 460(1):48–55. https://doi.org/10.1016/j.abb.2007.01.024
Chaturvedi P, Mishra M, Akhtar N, Gupta P, Mishra P, Tuli R (2012) Sterol glycosyltransferases-identification of members of gene family and their role in stress in Withania somnifera. Mol Biol Rep 39(10):9755–9764. https://doi.org/10.1007/s11033-012-1841-3
Liu YN, Hong LL, Liu M, Guo QC, Kong JQ (2021) Glycodiversifying testosterone with a promiscuous glycosyltransferase OsSGT2 from Ornithogalum saundersiae. ACS Synth Biol 10(12):3583–3594. https://doi.org/10.1021/acssynbio.1c00532
Kohara A, Nakajima C, Hashimoto K, Ikenaga T, Tanaka H, Shoyama Y, Yoshida S, Muranaka T (2005) A novel glucosyltransferase involved in steroid saponin biosynthesis in Solanum aculeatissimum. Plant Mol Biol 57(2):225–239. https://doi.org/10.1007/s11103-004-7204-2
Song W, Zhang C, Wu J, Qi J, Hua X, Kang L, Yuan Q, Yuan J, Xue Z (2022) Characterization of three Paris polyphylla glycosyltransferases from different UGT families for steroid functionalization. ACS Synth Biol. https://doi.org/10.1021/acssynbio.2c00103
XU Wen-juan, LI Xian-en, SUN Peng, ZHOU Li-li, SUN Tong-yu, QI Jian-jun (2013) Studies on expression level of genes related to abscisic acid and gibberellic acid in stratified Paris polyphylla var yunnanensis seeds. Zhong Cao Yao 44(3):338–343
Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31(13):3381–3385. https://doi.org/10.1093/nar/gkg520
Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8(4):477–486. https://doi.org/10.1007/bf00228148
Stucky DF, Arpin JC, Schrick K (2015) Functional diversification of two UGT80 enzymes required for steryl glucoside synthesis in Arabidopsis. J Exp Bot 66(1):189–201. https://doi.org/10.1093/jxb/eru410
Liu M, Kong JQ (2018) The enzymatic biosynthesis of acylated steroidal glycosides and their cytotoxic activity. Acta Pharm Sin B 8(6):981–994. https://doi.org/10.1016/j.apsb.2018.04.006
Poppenberger B, Fujioka S, Soeno K, George GL, Vaistij FE, Hiranuma S, Seto H, Takatsuto S, Adam G, Yoshida S, Bowles D (2005) The UGT73C5 of Arabidopsis thaliana glucosylates brassinosteroids. Proc Natl Acad Sci USA 102(42):15253–15258. https://doi.org/10.1073/pnas.0504279102
Augustin JM, Drok S, Shinoda T, Sanmiya K, Nielsen JK, Khakimov B, Olsen CE, Hansen EH, Kuzina V, Ekstrom CT, Hauser T, Bak S (2012) UDP-glycosyltransferases from the UGT73C subfamily in Barbarea vulgaris catalyze sapogenin 3-O-glucosylation in saponin-mediated insect resistance. Plant Physiol 160(4):1881–1895. https://doi.org/10.1104/pp.112.202747
Tang JR, Chen G, Lu YC, Tang QY, Song WL, Lin Y, Li Y, Peng SF, Yang SC, Zhang GH, Hao B (2021) Identification of two UDP-glycosyltransferases involved in the main oleanane-type ginsenosides in Panax japonicus var. major. Planta 253(5):91. https://doi.org/10.1007/s00425-021-03617-0
Hua X, Song W, Wang K, Yin X, Hao C, Duan B, Xu Z, Su T, Xue Z (2022) Effective prediction of biosynthetic pathway genes involved in bioactive polyphyllins in Paris polyphylla. Commun Biol 5(1):50. https://doi.org/10.1038/s42003-022-03000-z
Tapondjou LA, Ponou KB, Teponno RB, Mbiantcha M, Djoukeng JD, Nguelefack TB, Watcho P, Cadenas AG, Park HJ (2008) In vivo anti-inflammatory effect of a new steroidal saponin, mannioside A, and its derivatives isolated from Dracaena mannii. Arch Pharm Res 31(5):653–658. https://doi.org/10.1007/s12272-001-1208-3
Li J, Lv M, Du L, Yunga A, Hao S, Zhang Y, Zhang X, Guo L, Gao X, Deng L, Zhang X, Shi C, Guo F, Liu R, Fang B, Su Q, Hu X, Su X, Lin L, Liu Q, Wang Y, Qin Y, Zhang W, Li S, Liu C, Li H (2020) An enormous Paris polyphylla genome sheds light on genome size evolution and polyphyllin biogenesis. BioRxiv. https://doi.org/10.1101/2020.06.01.126920
Ma B, Liu X, Lu Y, Ma X, Wu X, Wang X, Jia M, Su P, Tong Y, Guan H, Jiang Z, Gao J, Huang L, Gao W (2019) A specific UDP-glucosyltransferase catalyzes the formation of triptophenolide glucoside from Tripterygium wilfordii Hook. f. Phytochemistry 166:112062. https://doi.org/10.1016/j.phytochem.2019.112062
Xu YL, Kong JQ (2020) OcUGT1-catalyzing glycodiversification of steroids through glucosylation and transglucosylation actions. Molecules. https://doi.org/10.3390/molecules25030475
Hoang NH, Hong SY, Huong NL, Park JW (2016) Biochemical characterization of recombinant UDP-glucose: sterol 3-O-glycosyltransferase from Micromonospora rhodorangea ATCC 31603 and enzymatic biosynthesis of sterol-3-O-beta-glucosides. J Microbiol Biotechnol 26(3):477–482. https://doi.org/10.4014/jmb.1511.11003
Srivastava P, Garg A, Misra RC, Chanotiya CS, Ghosh S (2021) UGT86C11 is a novel plant UDP-glycosyltransferase involved in labdane diterpene biosynthesis. J Biol Chem 297(3):101045. https://doi.org/10.1016/j.jbc.2021.101045
Sun G, Putkaradze N, Bohnacker S, Jonczyk R, Fida T, Hoffmann T, Bernhardt R, Hartl K, Schwab W (2020) Six uridine-diphosphate glycosyltransferases catalyze the glycosylation of bioactive C(13)-apocarotenols. Plant Physiol 184(4):1744–1761. https://doi.org/10.1104/pp.20.00953
Irmisch S, Jo S, Roach CR, Jancsik S, Man Saint Yuen M, Madilao LL, O’Neil-Johnson M, Williams R, Withers SG, Bohlmann J (2018) Discovery of UDP-glycosyltransferases and BAHD-acyltransferases involved in the biosynthesis of the antidiabetic plant metabolite montbretin A. Plant Cell 30(8):1864–1886. https://doi.org/10.1105/tpc.18.00406
Erthmann PO, Agerbirk N, Bak S (2018) A tandem array of UDP-glycosyltransferases from the UGT73C subfamily glycosylate sapogenins, forming a spectrum of mono- and bisdesmosidic saponins. Plant Mol Biol 97(1–2):37–55. https://doi.org/10.1007/s11103-018-0723-z
Shao H, He X, Achnine L, Blount JW, Dixon RA, Wang X (2005) Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula. Plant Cell 17(11):3141–3154. https://doi.org/10.1105/tpc.105.035055
Dai L, Qin L, Hu Y, Huang JW, Hu Z, Min J, Sun Y, Guo RT (2021) Structural dissection of unnatural ginsenoside-biosynthetic UDP-glycosyltransferase Bs-YjiC from Bacillus subtilis for substrate promiscuity. Biochem Biophys Res Commun 534:73–78. https://doi.org/10.1016/j.bbrc.2020.11.104
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 81974515)
Funding
This study was funded by No. 81974515.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interest
The authors declare that they have no competing interests.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
He, M., Guo, S., Yin, Y. et al. A novel sterol glycosyltransferase catalyses steroidal sapogenin 3-O glucosylation from Paris polyphylla var. yunnanensis. Mol Biol Rep 50, 2137–2146 (2023). https://doi.org/10.1007/s11033-022-08199-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-08199-y