Abstract
Background
Lilium genus consists of approximately 100 species and numerous varieties, many of which are interspecific hybrids, which result in a complicated genetic background. The germplasm identification, genetic relationship analysis, and breeding of Lilium rely on exploiting genetic information among different accessions. Hence, an attempt was made to develop new EST-SSR markers and study the molecular divergence among 65 genotypes of Lilium.
Methods and results
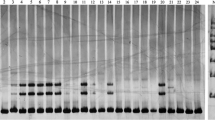
A total of 5509 EST-SSRs were identified from the high-throughput sequencing database of L. ‘Elodie’. After primer screening, six primer pairs with the most abundant polymorphic bands were selected from 100 primer pairs. Combined with the other 10 reported SSR primers, a total of 16 pairs detected genetic information with an average PIC value of 0.7583. The number of alleles per locus varied from four to 33, the expected heterozygosity varied from 0.3289 to 0.9231, and the observed heterozygosity varied from 0.2857 to 0.5122. Based on the phylogenic results, 22 Asiatic hybrids (A), seven Longiflorum × Asiatic hybrids (LA), as well as two native species were grouped. Eighteen Oriental hybrids (O) and nine Oriental × Trumpet (OT) hybrids, four native species, one LO, and one LL (L. pardalinum × L. longiflorum) variety were grouped.
Conclusions
Two major clusters were reported and a large number of genotypes were grouped based on UPGMA and STRUCTURE analysis methods. The PIC value as well as other parameters revealed that the EST-SSR markers selected were informative. In addition, the clustering pattern displayed better agreement with the cultivar’s pedigree. The newly identified SSRs in this study provides molecular markers for germplasm characterization and genetic diversity for Lilium.


Similar content being viewed by others
References
Long YY, Zhang JZ, Zhang LN (1999) Lily-the king of corm flowers. Jindun Press, Beijing, pp 1–2
Van Tuyl JM, Arens P, Shahin A, Marasek-Ciołakowska A, Barba-Gonzalez R, Kim HT, Lim KB (2018) Lilium. Ornamental crops. Handbook Plant Breeding 11:481–512
Comber HF (1949) A new classification of the genus Lilium. Lily Yearbook. R Hort Soc 13:86–105
Shahin A, Smulders MJM, van Tuyl JM, Arens P, Bakker FT (2014) Using multi-locus allelic sequence data to estimate genetic divergence among four Lilium (Liliaceae) cultivars. Front Plant Sci 5:567
Kong Y, Bai J, Lang L, Bao F, Dou X, Wang H, Shang H (2017) Floral scents produced by Lilium and Cardiocrinum species native to China. Biochem Syst Ecol 70:222–229
Lee CS, Kim SC, Yeau SH, Lee NS (2011) Major lineages of the genus Lilium (Liliaceae) based on nrDNA ITS sequences, with special emphasis on the Korean species. J Plant Biol 54:159–171
Kong Y, Wang H, Lang L, Dou X, Bai J (2021) Metabolome-based discrimination analysis of five Lilium bulbs associated with differences in secondary metabolites. Molecules 26:1340
Munafo JP, Gianfagna TJ (2011) Quantitative analysis of steroidal glycosides in different organs of Easter lily (Lilium longiflorum Thunb.) by LC-MS/MS. J Agric Food Chem 59:995–1004
Francis JA, Rumbeiha W, Nair MG (2004) Constituents in Easter lily flowers with medicinal activity. Life Sci 76(6):671–683
Kumar SPJ, Susmita C, Sripathy KV, Agarwal DK, Pal G, Singh AN, Kumar S, Rai AK, Simal–Gandara J (2022) Molecular characterization and genetic diversity studies of Indian soybean (Glycine max (L.) Merr.) cultivars using SSR markers. Mol Biol Rep 49:2129–2140
Ma L, Wang X, Yan M, Liu F, Zhang S, Wang X (2022) Genome survey sequencing of common vetch (Vicia sativa L.) and genetic diversity analysis of Chinese germplasm with genomic SSR markers. Mol Biol Rep 49:313–320
Nyabera LA, Nzuki IW, Runo SM, Amwayi PW (2021) Assessment of genetic diversity of pumpkins (Cucurbita spp.) from western Kenya using SSR molecular markers. Mol Biol Rep 48:2253–2260
Liu C, Dou Y, Guan X, Fu Q, Zhang Z, Hu Z, Zheng J, Lu Y, Li W (2017) De novo transcriptomic analysis and development of EST-SSRs for Sorbus pohuashanensis (Hance) Hedl. PLoS ONE 12(6):e0179219
Zietkiewicz E, Rafalski A, Labuda D (1994) Genome fingerprinting by simple sequence repeat (SSR) anchored polymerase chain reaction amplification. Genomics 20(2):176–183
Ukoskit K, Posudsavang G, Pongsiripat N, Chatwachirawong P, Klomsaard P, Poomipant P, Tragoonrung S (2018) Detection and validation of EST-SSR markers associated with sugar-related traits in sugarcane using linkage and association mapping. Genomics 111:1–9
Du F, Jiang J, Jia H, Zhao X, Wang W, Gao Q, Mao W, Wu Y, Zhang L, Grierson D, Xia Y, Gao Z (2015) Selection of generally applicable SSR markers for evaluation of genetic diversity and identity in Lilium. Biochem Syst Ecol 61:278–285
Lee SI, Park KC, Song YS, Son JH, Kwon SJ, Na JK, Kim JH, Kim NS (2011) Development of expressed sequence tag derived-simple sequence repeats in the genus Lilium. Genes Genom 33:727–733
Yuan S, Ge L, Liu C, Ming J (2013) The development of EST-SSR markers in Lilium regale and their cross-amplification in related species. Euphytica 189:393–419
Shahin A, van Kaauwen M, Esselink D, Bargsten JW, van Tuyl JM, Visser RG, Arens P (2012) Generation and analysis of expressed sequence tags in the extreme large genomes Lilium and Tulipa. BMC Genomics 13:640
Chen M, Nie G, Yang L, Zhang Y, Cai Y (2021) Homeotic transformation from stamen to petal in Lilium is associated with MADS-box genes and hormone signal transduction. Plant Growth Regul 95:49–64
Thiel T, Michalek W, Varshney RK, Graner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106:411–422
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386
Du F, Wu Y, Zhang L, Li XW, Zhao XY, Wang WH, Gao ZS, Xia YP (2015) De novo assembled transcriptome analysis and SSR marker development of a mixture of six tissues from Lilium Oriental hybrid ‘Sorbonne’. Plant Mol Biol Rep 33:281–293
Glaubitz JC (2004) CONVERT: a user friendly program to reformat diploid genotypic data for commonly used population genetic software packages. Mol Ecol Notes 4:309–310
Yeh FC, Boyle TJB (1997) Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg J Bot 129:157–163
Langella O (2007) Populations 1.2.30: population genetic software (individuals or populations distances, phylogenetic trees). France. http://bioinformatics.org/~tryphon/populations/. Accessed 14 Sept 2012
Letunic I, Bork P (2019) Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acid Res 47:W256–W259
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620)
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Kalia RK, Rai MK, Kalia S, Singh R, Dhawan AK (2011) Microsatellite markers: an overview of the recent progress in plants. Euphytica 177:309–334
Ma JQ, Ma CL, Yao MZ, Jin JQ, Wang ZL, Wang XC, Chen L (2012) Microsatellite markers from tea plant expressed sequence tags (ESTs) and their applicability for cross-species/genera amplification and genetic mapping. Sci Hortic 134:167–175
Qiu L, Yang C, Tian B, Yang JB, Liu A (2010) Exploiting EST databases for the development and characterization of EST-SSR markers in castor bean (Ricinus communis L.). BMC Plant Biol 10:278–287
Aggarwal RK, Hendre PS, Varshney RK, Bhat PR, Krishnakumar V, Singh L (2007) Identification, characterization and utilization of EST-derived genic microsatellite markers for genome analyses of coffee and related species. Theor Appl Genet 114:359–372
Baisakh N, Subudhi PK, Arumuganathan K, Parco AP, Harrison SA, Knott CA, Materne MD (2009) Development and interspecific transferability of genic microsatellite markers in Spartina spp. with different genome size. Aquat Bot 91:262–266
Varshney RK, Thiel T, Stein N, Langridge P, Graner A (2002) In silico analysis on frequency and distribution of microsatellites in ESTs of some cereal species. Cell Mol Biol Lett 7:537–546
Wang YW, Samuels TD, Wu YQ (2011) Development of 1030 genomic SSR markers in switchgrass. Theor Appl Genet 122:677–686
Wang Z, Yu G, Shi B, Wang X, Qiang H, Gao H (2014) Development and characterization of simple sequence repeat (SSR) markers based on RNA-sequencing of Medicago sativa and in silico mapping onto the M. truncatula genome. PLoS ONE 9(3):e92029
Tantasawat P, Trongchuen J, Prajongjai T, Seehalak W, Jittayasothorn Y (2011) Variety identification and comparative analysis of genetic diversity in yardlong bean (Vigna unguiculata spp. sesquipedalis) using morphological characters, SSR and ISSR analysis. Sci Hort 124:204–216
Lim KB, Barba-Gonzalez R, Zhou S, Ramanna MS, van Tuyl JM (2008) Interspecific hybridization in lily (Lilium): taxonomy and commercial aspects of using species hybrids in breeding. Floriculture, ornamental and plant biotechnology, vol 5. Global Science Books, Japan, pp 146–151
McRae EA (1998) Lilies: a guide for growers and collectors. Timber Press, Portland
Lim KB, Van Tuyl JM (2006) Lily, Lilium hybrids. Flower Breed Genet 19:517–537
Asano Y (1989) Lilium L. In: Tsukumoto Y (ed) The great dictionary of horticulture, vol 5. Sykakukum, Tokyo, pp 198–209
Funding
This work was supported by the Shanghai Agriculture Applied Technology Development Program, China (Grant No. X2020-02-08-00-12-F01463).
Author information
Authors and Affiliations
Contributions
MC and YZ conceived the theme of the study and designed the experiment; MC, GN and XL performed the experiment; MC, LY and YC analyzed the SSR data; MC wrote the manuscript. All co-authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethics approval
The article does not contain any studies with human participants or animals performed by any of the authors.
Consent for publication
All the authors have read and consented to submit the manuscript.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, M., Nie, G., Li, X. et al. Development of EST-SSR markers based on transcriptome sequencing for germplasm evaluation of 65 lilies (Lilium). Mol Biol Rep 50, 3259–3269 (2023). https://doi.org/10.1007/s11033-022-08083-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-08083-9