Abstract
Background
Biotechnologists seeking to develop marker-free transgenic plants have established co-transformation methods. For co-transformation using mixed Agrobacterium strains, the mix ratio of Agrobacterium strains and selection scheme may influence co-transformation frequency. This study used fluorescent GFP and RFP markers to compose different selection schemes for observation of the selective dynamics of transformed rice cells and to investigate the factors affecting co-transformation efficiency.
Methods and results
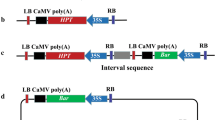
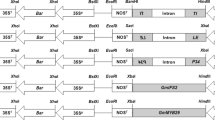
We utilized GFP and RFP markers in co-transformation and tested the combinations of an antibiotic-selectable vector (pGFP-HPT) and a single RFP vector (pRFP) and of two antibiotic-selectable vectors (pGFP-HPT and pRFP-HPT) in rice. The pGFP-HPT/pRFP combination resulted in 70.9% to 81.2% of co-transformation frequencies while lower frequencies (56.6% on average) were obtained with the pGFP-HPT/pRFP-HPT combination. Based on GFP/RFP segregation patterns, 55% of the pGFP-HPT/pRFP co-transformants contained unlinked T-DNAs and segregated single RFP progeny, which simulated the selection process of marker-free transgenic plants that carry an actual gene of interest. Transgene expression levels in the rice lines varied as revealed by RT-PCR, and tandem-linked T-DNAs were detected in co-transformants, suggesting that transgene expression might be affected by duplicated T-DNA structures.
Conclusion
Co-transformation via mixed Agrobacterium strains is feasible, and approximately 55% of the pGFP-HPT/pRFP co-transformants contained unlinked T-DNAs and segregated single RFP progeny. The pGFP-HPT/pRFP and the pGFP-HPT/pRFP-HPT vector combinations showed distinctive selective dynamics of transformed rice cells, suggesting that co-transformation efficiency depends on both vector system and selection scheme.



Similar content being viewed by others
Abbreviations
- GFP:
-
Green fluorescent protein
- RFP:
-
Red fluorescent protein
- HPT:
-
Hygromycin phosphotransferase
- Hyg:
-
Hygromycin
References
Liu F, Wang P, Xiong X, Fu P, Gao H, Ding X, Wu G (2020) Comparison of three Agrobacterium-mediated co-transformation methods for generating marker-free transgenic Brassica napus plants. Plant Methods 16:81. https://doi.org/10.1186/s13007-020-00628-y
Kapusi E, Hensel G, Coronado MJ, Broeders S, Marthe C, Otto I, Kumlehn J (2013) The elimination of a selectable marker gene in the doubled haploid progeny of co-transformed barley plants. Plant Mol Biol 81:149–160
Chen L, Marmey P, Taylor NJ, Brizard JP, Espinoza C, D′cruz P, Huet H, Zhang S, De Kochko A, Beachy RN, Fauquet CM (1998) Expression and inheritance of multiple transgenes in rice plants. Nat Biotechnol 16:1060–1064
Sripriya R, Raghupathy V, Veluthambi K (2008) Generation of selectable marker-free sheath blight resistant transgenic rice plants by efficient co-transformation of a cointegrate vector T-DNA and a binary vector T-DNA in one Agrobacterium tumefaciens strain. Plant Cell Rep 27:1635–1644
Komari T, Hiei Y, Saito Y, Murai N, Kumashiro T (1996) Vectors carrying two separate T-DNAs for co-transformation of higher plants mediated by Agrobacterium tumefaciens and segregation of transformants free from selection markers. Plant J 10:165–174
Breitler JC, Meynard D, Van Boxtel J, Royer M, Bonnot F, Cambillau L, Guiderdoni E (2004) A novel two T-DNA binary vector allows efficient generation of marker-free transgenic plants in three elite cultivars of rice (Oryza sativa L.). Transgenic Res 13:271–287
De Buck S, Jacobs A, Van Montagu M, Depicker A (1998) Agrobacterium tumefaciens transformation and cotransformation frequencies of Arabidopsis thaliana root explants and tobacco protoplasts. Mol Plant Microbe Interact 11:449–457
Park J, Lee YK, Kang BK, Chung WI (2004) Co-transformation using a negative selectable marker gene for the production of selectable marker gene-free transgenic plants. Theor Appl Genet 109:1562–1567
De Buck S, Podevin N, Nolf J, Jacobs A, Depicker A (2009) The T-DNA integration pattern in Arabidopsis transformants is highly determined by the transformed target cell. Plant J 60:134–145
De Neve M, De Buck S, Jacobs A, Van Montagu M, Depicker A (1997) T-DNA integration patterns in co-transformed plant cells suggest that T-DNA repeats originate from co-integration of separate T-DNAs. Plant J 11:15–29
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6:271–282
Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H (2006) Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J 47:969–976
De Buck S, Van Montagu M, Depicker A (2001) Transgene silencing of invertedly repeated transgenes is released upon deletion of one of the transgenes involved. Plant Mol Biol 46:433–445
Acknowledgements
The authors are grateful to Xiaowei Zou, Xiaoqing Gao, Haihe Yang, Haiyan Qu, and Suqin Zheng for their experimental assistance.
Funding
This work was financially supported by the State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products in Zhejiang Academy of Agricultural Sciences, and by the National Natural Science Foundation of China (Grant No. 31672016).
Author information
Authors and Affiliations
Contributions
LL, XT, LW, JZ, JZ and HH conducted the experiments, collected and analyzed the data. LL and SQ wrote the manuscript. SQ and DL designed the research schemes and supervised the whole process. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to publication
The authors consent to publish all the data in this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, L., Tian, X., Wang, L. et al. Characterization of Agrobacterium-mediated co-transformation events in rice using green and red fluorescent proteins. Mol Biol Rep 49, 9613–9622 (2022). https://doi.org/10.1007/s11033-022-07864-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07864-6