Abstract
Background
Chilli is an important commercial crop with positive returns tendency. Phytophthora root rot causes drastic damage to chilli plant. Dearth of detecting marker trait associations is a major hinderance in practicing marker assisted selection in chilli breeding.
Methods and results
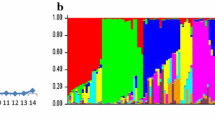
Herein, 110 chilli accessions were assessed for 15 agronomic traits under control and disease infected conditions for two crop seasons (2018–2019). The SSR genotyping revealed high values of major allele frequency (MAF = 0.70), genetic diversity (GD = 0.39) and Polymorphic Information Content (PIC = 0.31). Principal coordinate analysis and population structure analysis showed distribution of diverse genotypes in all groups by dividing 110 genotypes in three populations and nine sub-populations. The UPGMA based Archaeopteryx tree was in concordance with population structure analysis. Linkage disequilibrium analysis evaluated that LD decays within 3–10 bp. Marker trait association (MTA) revealed the associations of 35 SSRs with 14 morphological traits. The significant MTA for marker CAeMS073 with relative leaf damage (RLD, 0.183 R2) under control and treated conditions was consistently observed in both models. The markers, CAMS173 and CAMS194 were found to be strongly associated with RLD and Disease Index (DI), respectively. The absence of MTA was detected for height of first branch.
Conclusion
The MTAs reported in this study can facilitate marker assisted breeding for developing chilli germplasm resistant against Phytophthora capsici.



Similar content being viewed by others
References
Fahrurrozi F, Ganefianti DW (2019) Early yield responses of three promising chili pepper hybrids to different mulch types. Asian J Agric Biol 7:548–554
Ahmad A, Aslam MN, Qurashi F, Ashraf W, Raheel M, Shakeel Q, Saleem K (2020) Molecular diversity and phylogenetic reconstruction of Pepper mild mottle virus isolates from Pakistan. Asian J Agric Biol. https://doi.org/10.35495/ajab.2020.09.464
Barchenger DW, Lamour KH, Bosland PW (2018) Challenges and strategies for breeding resistance in Capsicum annuum to the multifarious pathogen, Phytophthora capsici. Front Plant Sci 9:628
Hausbeck MK, Lamour KH (2004) Phytophthora capsici on vegetable crops: research progress and management challenges. Plant Dis 88:1292–1303
Zhang HX, Ali M, Feng XH, Jin JH, Huang LJ, Khan A, Lv JG, Gao SY, Luo DX, Gong ZH (2019) A novel transcription factor CaSBP12 gene negatively regulates the defense response against Phytophthora capsici in pepper (Capsicum annuum L.). Int J Mol Sci 20:48
Chunthawodtiporn J, Hill T, Stoffel K, VanDeynze A (2018) Quantitative trait loci controlling fruit size and other horticultural traits in bell pepper (Capsicum annuum). Plant Genome 11:160125
Thabuis A, Lefebvre V, Bernard G, Daubeze AM, Phaly T, Pochard E, Palloix A (2004) Phenotypic and molecular evaluation of a recurrent selection program for a polygenic resistance to Phytophthora capsici in pepper. Theor Appl Genet 109:342–351
Kalu SE, Ubi GM, Osuagwu AN, Ekpo IA, Edem LU (2021) Microsatellite fingerprinting, enzymes activity and chlorophyll profiling of local lines of air potato yam (Dioscorea bulbifera L.) for salt tolerance. Asian J Agric Biol 1–14
Ibiza VP, Blanca J, Cañizares J, Nuez F (2012) Taxonomy and genetic diversity of domesticated Capsicum species in the Andean region. Genet Resour Crop Evol 59:1077–1088
Rabuma T, Gupta OP, Chhokar V (2020) Phenotypic characterization of chili pepper (Capsicum annuum L.) under Phytophthora capsici infection and analysis of genetic diversity among identified resistance accessions using SSR markers. Physiol Mol Plant Pathol 112:101539
Bosland PW, Lindsey DL (1991) A seedling screen for Phytophthora root rot of pepper, Capsicum annuum. Plant Dis 75:1048–1050
Aklilu S, Ayana G, Abebie B, Abdissa T (2018) Screening for resistance sources in local and exotic hot pepper genotypes to Fusarium wilt (Fusarium oxysporium) and associated quality traits in Ethiopia. Adv Crop Sci Technol 6:367–376
Widmer TL, Graham JH, Mitchell DJ (1998) Histological comparison of fibrous root infection of disease-tolerant and susceptible citrus hosts by Phytophthora nicotianae and P. palmivora. Phytopathology 88:389–395
Hameed M, Keitel C, Ahmad N, Mahmood T, Trethowan R (2015) Screening of tomatoes germplasm for heat stress tolerance under controlled conditions. Procedia Environ Sci 29:173–174
Daudi A, O’Brien JA (2012) Detection of hydrogen peroxide by DAB staining in Arabidopsis leaves. Bio-Protoc 2:263–263
Ruley AT, Sharma NC, Sahi SV (2004) Antioxidant defense in a lead accumulating plant, Sesbania drummondii. Plant Physiol Biochem 42:899–906
Doyle JJ (1990) Isolation of plant DNA from fresh tissue. Focus (Madison) 12:13–15
McGregor C, Waters V, Nambeesan S, MacLean D, Candole BL, Conner P (2011) Genotypic and phenotypic variation among pepper accessions resistant to Phytophthora capsici. HortScience 46:1235–1240
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Earl DA (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Monostori I, Szira F, Tondelli A, Arendas T, Gierczik K, Cattivelli L, Galiba G, Vagujfalvi A (2017) Genome-wide association study and genetic diversity analysis on nitrogen use efficiency in a Central European winter wheat (Triticum aestivum L.) collection. PLoS ONE 12:e0189265
Steel RG, Torrie JH (1986) Principles and procedures of statistics: a biometrical approach. McGraw Hill, New York
Federer WT, Raghavarao D (1975) On augmented designs. Biometrics 31:29–35
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21(9):2128–2129
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Dhaliwal MS, Yadav A, Jindal SK (2014) Molecular characterization and diversity analysis in chilli pepper using simple sequence repeats (SSR) markers. Afr J Biotechnol 13:31
Rahevar PM, Patel JN, Kumar S, Acharya RR (2019) Morphological, biochemical and molecular characterization for genetic variability analysis of Capsicum annuum. Vegetos 32:131–141
Pacheco OA, Hernández VS, Rocha RV, González RA, Oyama K (2012) Genetic diversity and structure of pepper (Capsicum annuum L.) from Northwestern Mexico analyzed by microsatellite markers. Crop Sci 52:231–241
Oh SJ, Song JY, Lee JR, Lee GA, Ko HC, Stoilova T, Krasteva L, Kim YG, Rhee JH, Gwag JG, Ro NY (2012) Evaluation of genetic diversity of red pepper landraces (Capsicum annuum L.) from Bulgaria using SSR markers. J Korean Soc Int Agric 24:547–556
Meng C, Wei X, Zhao Y, Yuan Y, Yang S, Wang Z, Zhang X, Sun J, Zheng X, Yao Q, Zhang Q (2017) Genetic diversity analysis of Capsicum genus by SSR markers. Mol Plant Breed. https://doi.org/10.5376/mpb.2017.08.008
Sharmin A, Hoque ME, Haque MM, Khatun F (2018) Molecular diversity analysis of some chilli (Capsicum spp.) genotypes using SSR markers. Am J Plant Sci 9:368–379
Khan MMH, Rafii MY, Ramlee SI, Jusoh M, Al Mamun M, Halidu J (2021) DNA fingerprinting, fixation-index (Fst), and admixture mapping of selected Bambara groundnut (Vigna subterranea [L.] Verdc.) accessions using ISSR markers system. Sci Rep 11:1–23
Yu J, Zhao H, Zhu T, Chen L, Peng J (2013) Transferability of rice SSR markers to Miscanthus sinensis, a potential biofuel crop. Euphytica 191:455–468
Loaiza FF, Ritland K, Cancino JAL, Tanksley SD (1989) Patterns of genetic variation of the genus Capsicum (Solanaceae) in Mexico. Plant Syst Evol 165:159–188
Poczai P, Varga I, Bell NE, Hyvönen J (2011) Genetic diversity assessment of bittersweet (Solanum dulcamara, Solanaceae) germplasm using conserved DNA-derived polymorphism and intron-targeting markers. Ann Appl Biol 159:141–153
Frankham R, Ballou SEJD, Briscoe DA, Ballou JD (2002) Introduction to conservation genetics. Cambridge University Press, Cambridge
N’Goran JAK, Laurent V, Risterucci A-M, Lanaud C (2000) The genetic structure of cocoa populations (Theobroma cacao L.) revealed by RFLP analysis. Euphytica 115:83–90
Sadiyah H, Ashari S, Waluyo B, Soegianto A (2021) Genetic diversity and relationship of husk tomato (Physalis spp.) from East Java Province revealed by SSR markers. Biodiversitas 22:184–192
Ragsdale AP, Gravel S (2020) Unbiased estimation of linkage disequilibrium from unphased data. Mol Biol Evol 37:923–932
Slatkin M (1994) Linkage disequilibrium in growing and stable populations. Genetics 137:331–336
Taranto F, D’Agostino N, Greco B, Cardi T, Tripodi P (2016) Genome-wide SNP discovery and population structure analysis in pepper (Capsicum annuum) using genotyping by sequencing. BMC Genomics 17:1–13
Fayaz F, Sarbarzeh MA, Talebi R, Azadi A (2019) Genetic diversity and molecular characterization of Iranian durum wheat landraces (Triticum turgidum durum (Desf.) Husn.) using DArT markers. Biochem Genet 57:98–116
Moreira AFP, Ruas PM, de Fátima Ruas C, Baba VY, Giordani W, Arruda IM, Rodrigues R, Gonçalves LSA (2018) Genetic diversity, population structure and genetic parameters of fruit traits in Capsicum chinense. Sci Hortic (Amsterdam) 236:1–9
Esyanti RR, Dwivany FM, Mahani S, Nugrahapraja H, Meitha K (2019) Foliar application of chitosan enhances growth and modulates expression of defense genes in chilli pepper (Capsicum annuum L.). Aust J Crop Sci 13:55–60
Bagga S, Lucero Y, Apodaca K, Rajapakse W, Lujan P, Ortega JL, Sengupta-Gopalan C (2019) Chile (Capsicum annuum) plants transformed with the RB gene from Solanum bulbocastanum are resistant to Phytophthora capsici. PLoS ONE 14:0223213
Ghorbanipour A, Rabiei B, Rahmanpour S, Khodaparast SA (2019) Association analysis of charcoal rot disease resistance in soybean plant. Pathol J 35:189
Hittalmani S, Huang N, Courtois B, Venuprasad R, Shashidhar HE, Zhuang JY, Zheng KL, Liu GF, Wang GC, Sidhu JS, Srivantaneeyakul S (2003) Identification of QTL for growth-and grain yield-related traits in rice across nine locations of Asia. Theor Appl Genet 107:679–690
Acknowledgements
The authors acknowledge the support under the Higher Education Commission NRPU Project 6137 (2016) titled “Development of double Haploid Lines of Chilies Resistant to Phytophthora Root Rot”.
Funding
This study was funded by Higher Education Commission, Pakistan (Grant No. 6137/Punjab/NRPU/R&D/HEC/2016).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with animals or human performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bukhari, T., Rana, R.M., Hassan, M.U. et al. Genetic diversity and marker trait association for phytophthora resistance in chilli. Mol Biol Rep 49, 5717–5728 (2022). https://doi.org/10.1007/s11033-022-07635-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07635-3