Abstract
Background
Obesity is a complex genetic-based pediatric disorder which triggers life-threatening conditions. Therefore, the understanding the molecular mechanisms of obesity has been a significant approach in medicine. Computational methods allow rapid and comprehensive pathway analysis, which is important for generation of diagnosis and treatment of obesity.
Methods and results
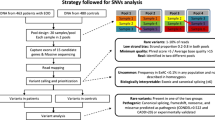
Aims of our study are to comprehensively investigate genetic characteristics of obesity in children with non-syndromic, early-onset (< 7 years), and severe obesity (BMI-SDS > 3) through computational approaches. First, the mutational analyses of 41 of obesity-related genes in 126 children with non-syndromic early-onset severe obesity and 76 healthy non-obese controls were performed using the next generation sequencing (NGS) technique, and the NGS data analyzed by using bioinformatics methods. Then, the relationship between pathogenic variants and anthropometric/biochemical parameters was further evaluated. Obtained results demonstrated that the 15 genes (ADIPOQ, ADRB2, ADRB3, IRS1, LEPR, NPY, POMC, PPARG, PPARGC1A, PPARGC1B, PTPN1, SLC22A1, SLC2A4, SREBF1 and UCP1) which directly related to obesity found linked together via biological pathways and/or functions. Among these genes, IRS1, PPARGC1A, and SLC2A4 stand out as the most central ones. Furthermore, 12 of non-synonymous pathogenic variants, including six novels, were detected on ADIPOQ (G90S and D242G), ADRB2 (V87M), PPARGC1A (E680G, A477T, and R656H), UCP1 (Q44R), and IRS1 (R302Q, R301H, R301C, H250P, and H250N) genes.
Conclusion
We propose that 12 of non-synonymous pathogenic variations detected on ADIPOQ, ADRB2, PPARGC1A, UCP1, and IRS1 genes might have a cumulative effect on the development and progression of obesity.



Similar content being viewed by others
Data availability
All data provided as supplementary files.
Code availability
All software and packages used in this study can be seen in the methodology section. The required information to install and run these tools can be obtained from the provided references.
References
Kansra AR, Lakkunarajah S, Jay MS (2021) Childhood and adolescent obesity: a review. Front Pediatr 8:866. https://doi.org/10.3389/fped.2020.581461
Kaila B, Raman M (2008) Obesity: a review of pathogenesis and management strategies. Can J Gastroenterol 22:61–68. https://doi.org/10.1155/2008/609039
Blüher M (2019) Obesity: global epidemiology and pathogenesis. Nat Rev Endocrinol 15:288–298. https://doi.org/10.1038/s41574-019-0176-8
Goodarzi MO (2018) Genetics of obesity: what genetic association studies have taught us about the biology of obesity and its complications. Lancet Diabetes Endocrinol 6:223–236. https://doi.org/10.1016/S2213-8587(17)30200-0
Thaker VV (2017) Genetic and epigenetic causesof obesity. Adolesc Med State Art Rev 28:379–405
Müller MJ, Geisler C, Blundell J et al (2018) The case of GWAS of obesity: does body weight control play by the rules? Int J Obes 42:1395–1405. https://doi.org/10.1038/s41366-018-0081-6
Loos RJF, Yeo GSH (2021) The genetics of obesity: from discovery to biology. Nat Rev Genet. https://doi.org/10.1038/s41576-021-00414-z
Bouchard C (2021) Genetics of obesity: what we have learned over decades of research. Obesity (Silver Spring, Md) 29:802–820. https://doi.org/10.1002/oby.23116
Trier C, Hollensted M, Schnurr TM et al (2021) Obesity treatment effect in Danish children and adolescents carrying Melanocortin-4 receptor mutations. Int J Obes 45:66–76. https://doi.org/10.1038/s41366-020-00673-6
Santos JL, Cortés VA (2021) Eating behaviour in contrasting adiposity phenotypes: monogenic obesity and congenital generalized lipodystrophy. Obes Rev 22:e13114. https://doi.org/10.1111/obr.13114
da Fonseca ACP, Mastronardi C, Johar A et al (2017) Genetics of non-syndromic childhood obesity and the use of high-throughput DNA sequencing technologies. J Diabetes Complicat 31:1549–1561. https://doi.org/10.1016/j.jdiacomp.2017.04.026
Mason K, Page L, Balikcioglu PG (2014) Screening for hormonal, monogenic, and syndromic disorders in obese infants and children. Pediatr Ann 43:e218–e224. https://doi.org/10.3928/00904481-20140825-08
Singh RK, Kumar P, Mahalingam K (2017) Molecular genetics of human obesity: a comprehensive review. C R Biol 340:87–108. https://doi.org/10.1016/j.crvi.2016.11.007
Cheng M, Mei B, Zhou Q et al (2018) Computational analyses of obesity associated loci generated by genome-wide association studies. PLoS ONE 13:1–13. https://doi.org/10.1371/journal.pone.0199987
Chakraborty BM, Chakraborty R (2012) Bioinformatics of obesity. In: Chakraborty R, Rao CR, Sen PBT (eds) Handbook of statistics. Elsevier, Amsterdam, pp 433–477
Rohde K, Keller M, la Cour Poulsen L et al (2019) Genetics and epigenetics in obesity. Metabolism 92:37–50. https://doi.org/10.1016/j.metabol.2018.10.007
Neyzi O, Bundak R, Gökçay G et al (2015) Reference values for weight, height, head circumference, and body mass index in Turkish children. J Clin Res Pediatric Endocrinol 7:280–293. https://doi.org/10.4274/jcrpe.2183
Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38:e164–e164. https://doi.org/10.1093/nar/gkq603
Ng PC, Henikoff S (2003) SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res 31:3812–3814. https://doi.org/10.1093/nar/gkg509
Adzhubei I, Jordan DM, Sunyaev SR (2013) Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet 76:7.20.1-7.20.41. https://doi.org/10.1002/0471142905.hg0720s76
Rentzsch P, Witten D, Cooper GM et al (2019) CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res 47:D886–D894. https://doi.org/10.1093/nar/gky1016
Davydov EV, Goode DL, Sirota M et al (2010) Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLOS Comput Biol 6:e1001025
Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A (2010) Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res 20:110–121. https://doi.org/10.1101/gr.097857.109
Chun S, Fay JC (2009) Identification of deleterious mutations within three human genomes. Genome Res 19:1553–1561. https://doi.org/10.1101/gr.092619.109
Schwarz JM, Cooper DN, Schuelke M, Seelow D (2014) MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 11:361–362. https://doi.org/10.1038/nmeth.2890
Reva B, Antipin Y, Sander C (2011) Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res 39:e118–e118. https://doi.org/10.1093/nar/gkr407
Shihab HA, Gough J, Cooper DN et al (2013) Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat 34:57–65. https://doi.org/10.1002/humu.22225
Liu X, Wu C, Li C, Boerwinkle E (2016) dbNSFP v3.0: a one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum Mutat 37:235–241. https://doi.org/10.1002/humu.22932
Ioannidis NM, Rothstein JH, Pejaver V et al (2016) REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet 99:877–885. https://doi.org/10.1016/j.ajhg.2016.08.016
Fox J, Leanage A (2016) R and the journal of statistical software. J Stat Softw. https://doi.org/10.18637/jss.v073.i02
Mayakonda A, Lin D-C, Assenov Y et al (2018) Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res 28:1747–1756. https://doi.org/10.1101/gr.239244.118
Bindea G, Mlecnik B, Hackl H et al (2009) ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25:1091–1093. https://doi.org/10.1093/bioinformatics/btp101
Shannon P, Markiel A, Ozier O et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. https://doi.org/10.1101/gr.1239303
Kanehisa M, Furumichi M, Tanabe M et al (2017) KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45:D353–D361. https://doi.org/10.1093/nar/gkw1092
Wu C-C, Bratton SB (2013) Regulation of the intrinsic apoptosis pathway by reactive oxygen species. Antioxid Redox Signal 19:546–558. https://doi.org/10.1089/ars.2012.4905
Franzago M, Fraticelli F, Marchioni M et al (2021) Fat mass and obesity-associated (FTO) gene epigenetic modifications in gestational diabetes: new insights and possible pathophysiological connections. Acta Diabetol 58:997–1007. https://doi.org/10.1007/s00592-020-01668-5
Garfield AS, Lam DD, Marston OJ et al (2009) Role of central melanocortin pathways in energy homeostasis. Trends Endocrinol Metab 20:203–215. https://doi.org/10.1016/j.tem.2009.02.002
Cao H (2014) Adipocytokines in obesity and metabolic disease. J Endocrinol 220:T47-59. https://doi.org/10.1530/JOE-13-0339
Jeon S-M (2016) Regulation and function of AMPK in physiology and diseases. Exp Mol Med 48:e245. https://doi.org/10.1038/emm.2016.81
Salas-Pérez F, Ramos-Lopez O, Mansego ML et al (2019) DNA methylation in genes of longevity-regulating pathways: association with obesity and metabolic complications. Aging 11:1874–1899. https://doi.org/10.18632/aging.101882
Ye J (2013) Mechanisms of insulin resistance in obesity. Front Med 7:14–24. https://doi.org/10.1007/s11684-013-0262-6
Sharma M, Aggarwal S, Nayar U et al (2021) Differential expression of insulin receptor substrate-1(IRS-1) in visceral and subcutaneous adipose depots of morbidly obese subjects undergoing bariatric surgery in a tertiary care center in north India; SNP analysis and correlation with metabolic profile. Diabetes Metab Syndr 15:981–986. https://doi.org/10.1016/j.dsx.2021.04.014
Baroni MG, Arca M, Sentinelli F et al (2001) The G972R variant of the insulin receptor substrate-1 (IRS-1) gene, body fat distribution and insulin-resistance. Diabetologia 44:367–372. https://doi.org/10.1007/s001250051628
Le Fur S, Le Stunff C, Bougnères P (2002) Increased insulin resistance in obese children who have both 972 IRS-1 and 1057 IRS-2 polymorphisms. Diabetes 51(Suppl 3):S304–S307. https://doi.org/10.2337/diabetes.51.2007.s304
Menzaghi C, Trischitta V, Doria A (2007) Genetic influences of adiponectin on insulin resistance, type 2 diabetes, and cardiovascular disease. Diabetes 56:1198–1209. https://doi.org/10.2337/db06-0506
Palit SP, Patel R, Jadeja SD et al (2020) A genetic analysis identifies a haplotype at adiponectin locus: association with obesity and type 2 diabetes. Sci Rep 10:2904. https://doi.org/10.1038/s41598-020-59845-z
Kroll C, Mastroeni SSBS, Veugelers PJ, Mastroeni MF (2019) Associations of ADIPOQ and LEP gene variants with energy intake: a systematic review. Nutrients. https://doi.org/10.3390/nu11040750
Buzzetti R, Petrone A, Zavarella S et al (2007) The glucose clamp reveals an association between adiponectin gene polymorphisms and insulin sensitivity in obese subjects. Int J Obes 31:424–428. https://doi.org/10.1038/sj.ijo.0803419
Vozarova de Courten B, Hanson RL, Funahashi T et al (2005) Common polymorphisms in the adiponectin gene ACDC are not associated with diabetes in Pima Indians. Diabetes 54:284–289. https://doi.org/10.2337/diabetes.54.1.284
Kantartzis K, Fritsche A, Machicao F et al (2006) The -8503 G/A polymorphism of the adiponectin receptor 1 gene is associated with insulin sensitivity dependent on adiposity. Diabetes Care 29:464
Jungtrakoon P, Plengvidhya N, Tangjittipokin W et al (2011) Novel adiponectin variants identified in type 2 diabetic patients reveal multimerization and secretion defects. PLoS ONE 6:e26792. https://doi.org/10.1371/journal.pone.0026792
Hivert M-F, Manning AK, McAteer JB et al (2008) Common variants in the adiponectin gene (ADIPOQ) associated with plasma adiponectin levels, type 2 diabetes, and diabetes-related quantitative traits: the Framingham Offspring Study. Diabetes 57:3353–3359. https://doi.org/10.2337/db08-0700
Auger C, Kajimura S (2021) Detouring adrenergic stimulation to induce adipose thermogenesis. Nat Rev Endocrinol 17:579–580. https://doi.org/10.1038/s41574-021-00546-6
Prior SJ, Goldberg AP, Ryan AS (2011) ADRB2 haplotype is associated with glucose tolerance and insulin sensitivity in obese postmenopausal women. Obesity (Silver Spring, Md) 19:396–401. https://doi.org/10.1038/oby.2010.197
Mitra SR, Tan PY, Amini F (2019) Association of ADRB2 rs1042713 with obesity and obesity-related phenotypes and its interaction with dietary fat in modulating glycaemic indices in Malaysian adults. J Nutr Metab 2019:8718795. https://doi.org/10.1155/2019/8718795
Corbalán MS, Marti A, Forga L et al (2002) Beta(2)-adrenergic receptor mutation and abdominal obesity risk: effect modification by gender and HDL-cholesterol. Eur J Nutr 41:114–118. https://doi.org/10.1007/s00394-002-0363-5
Hellström L, Large V, Reynisdottir S et al (1999) The different effects of a Gln27Glu beta 2-adrenoceptor gene polymorphism on obesity in males and in females. J Intern Med 245:253–259. https://doi.org/10.1046/j.1365-2796.1999.0437e.x
Vega RB, Huss JM, Kelly DP (2000) The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol Cell Biol 20:1868–1876. https://doi.org/10.1128/MCB.20.5.1868-1876.2000
Tan L-J, Zhu H, He H et al (2014) Replication of 6 obesity genes in a meta-analysis of genome-wide association studies from diverse ancestries. PLoS ONE 9:e96149. https://doi.org/10.1371/journal.pone.0096149
Vimaleswaran KS, Radha V, Anjana M et al (2006) Effect of polymorphisms in the PPARGC1A gene on body fat in Asian Indians. Int J Obes 30:884–891. https://doi.org/10.1038/sj.ijo.0803228
Das SK (2007) Thr394Thr polymorphism (rs2970847) of PPARGC1A gene and obesity in Asian Indians. Int J Obes 31:562–563
Esterbauer H, Oberkofler H, Linnemayr V et al (2002) Peroxisome proliferator-activated receptor-gamma coactivator-1 gene locus: associations with obesity indices in middle-aged women. Diabetes 51:1281–1286. https://doi.org/10.2337/diabetes.51.4.1281
Jia J, Tian Y, Cao Z et al (2010) The polymorphisms of UCP1 genes associated with fat metabolism, obesity and diabetes. Mol Biol Rep 37:1513–1522. https://doi.org/10.1007/s11033-009-9550-2
Brondani LA, de Souza BM, Assmann TS et al (2014) Association of the UCP polymorphisms with susceptibility to obesity: case-control study and meta-analysis. Mol Biol Rep 41:5053–5067. https://doi.org/10.1007/s11033-014-3371-7
Mills EL, Harmon C, Jedrychowski MP et al (2021) UCP1 governs liver extracellular succinate and inflammatory pathogenesis. Nat Metab 3:604–617. https://doi.org/10.1038/s42255-021-00389-5
Ding B, Kull B, Liu Z et al (2005) Human neuropeptide Y signal peptide gain-of-function polymorphism is associated with increased body mass index: possible mode of function. Regul Pept 127:45–53. https://doi.org/10.1016/j.regpep.2004.10.011
Katus U, Villa I, Ringmets I et al (2021) Neuropeptide Y gene variants in obesity, dietary intake, blood pressure, lipid and glucose metabolism: a longitudinal birth cohort study. Peptides 139:170524. https://doi.org/10.1016/j.peptides.2021.170524
Acknowledgements
We thank Assoc. Prof., M.D., Bahri Evren, MD (Inonu University Medical Faculty, Endocrinology and Diabetes Department, Malatya, Turkey), Prof., MD. Ibrahim Sahin (Inonu University Medical Faculty, Endocrinology and Diabetes Department, Malatya, Turkey), MD. Yusuf Curek (Antalya Training and Research Hospital, Pediatric Endocrinology Department, Antalya, Turkey), Prof., MD. Aysun Bideci (Gazi University Medical Faculty, Pediatric Endocrinology, and Diabetes Department, Ankara, Turkey), Prof., MD. Ayla Guven (University of Health Sciences, Goztepe Training and Research Hospital, Pediatric Endocrinology Department, Istanbul, Turkey), Assoc. Prof., MD. Erdal Eren (Uludag University Medical Faculty, Pediatric Endocrinology and Diabetes Department, Bursa, Turkey), Assoc. Prof., MD. Ozlem Sangun (Baskent University Medical Faculty, Pediatric Endocrinology and Diabetes Department, Adana, Turkey), Assoc. Prof., MD. Atilla Cayir (Erzurum Training and Research Hospital, Pediatric Endocrinology Department, Erzurum, Turkey), Prof., MD. Pelin Bilir (Ankara University Medical Faculty, Pediatric Endocrinology, and Diabetes Department, Ankara, Turkey), Prof., MD. Ayca Torel Ergur (Ufuk University Medical Faculty, Pediatric Endocrinology, and Diabetes Department, Ankara, Turkey) and Prof., MD. Oya Ercan (Istanbul University Cerrahpasa Medical Faculty, Pediatric Endocrinology, and Diabetes, İstanbul, Turkey) for providing patient and/or control samples to this study.
Funding
This study was funded by Inonu University Scientific Research Centre (Grant Number 2018-20).
Author information
Authors and Affiliations
Contributions
AA took part in the design of the study, statistical analyses, evaluation of the results, and contributed to the writing of the manuscript. AK takes part in the design of the study, performed bioinformatics analyses, takes part in the evaluation of the results, generated the published figures, and contributed to the writing of the manuscript. AO takes part in the evaluating bioinformatics analysis and contributed to the writing and editing of the manuscript. SA and DT took part in evaluating the results and contributed to the writing of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
Ayşehan Akinci declares that she has no conflict of interest. Altan Kara declares that he has no conflict of interest. Aykut Özgür declares that he has no conflict of interest. Doğa Türkkahraman declares that he has no conflict of interest. Soner Aksu declares that he has no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Akinci, A., Kara, A., Özgür, A. et al. Genomic analysis to screen potential genes and mutations in children with non-syndromic early onset severe obesity: a multicentre study in Turkey. Mol Biol Rep 49, 1883–1893 (2022). https://doi.org/10.1007/s11033-021-06999-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06999-2