Abstract
Backgound
Bladder cancer (BCa) is a heterogeneous disease caused by the interaction between environmental and genetic risk factors. The goal of this case–control study was to evaluate the implication of a selected SNP panel in the risk of BCa development in a Tunisian cohort. We were also interested in studying the interaction between this predictive panel and environmental risk factors.
Methods
The case/control cohort was composed with 249 BCa cases and 255 controls. The designed Bladder cancer hereditary panel (BCHP) was composed of 139 selected variants. These variants were genotyped by an amplification-based targeted Next-Generation Sequencing (NGS) on the Ion Torrent Proton sequencer (Life Technologies, Ion Torrent technology).
Results
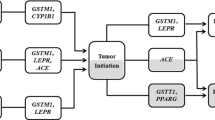
We have found that rs162555, rs2228000, rs10936599, rs710521, rs3752645, rs804276, rs4639, rs4881400 and rs288980 were significantly associated with decreased risk of bladder cancer. However the homozygous genotypes for VPS37C (rs7104333, A/A), MPG (rs1013358, C/C) genes or the heterozygous genotype for ARNT gene (rs1889740, rs2228099, rs2256355, rs2864873), GSTA4 (rs17614751) and APOBR/IL27 (rs17855750) were significantly associated with increased risk of bladder cancer development compared to reference group (OR 2.53, 2.34, 1.99, 2.00, 2.00, 1.47, 1.96 and 2.27 respectively). We have also found that non–smokers patients harboring heterozygous genotypes for ARNT/rs2864873 (A > G), ARNT/ rs1889740 (C > T) or GSTA4/rs17614751 (G–A) were respectively at 2.775, 3.069 and 6.608-fold increased risk of Bca development compared to non-smokers controls with wild genotypes. Moreover the ARNT CT (rs1889740), ARNT CG (rs2228099), ARNT TC (rs2864873) and GSS GA genotypes were associated with an increased risk of BCa even in absence of professional risk factors. Finally the decision-tree analysis produced a three major BCa classes. These three classes were essentially characterized by an intensity of tobacco use more than 20 pack years (PY) and the CYP1A2 (rs762551) genotype.
Conclusions
The determined association between environmental factors, genetic variations and the risk of Bca development may provide additional information to urologists that may help them for clinical assessment and treatment decisions. Nevertheless, the underlying mechanisms through which these genes or SNPs affect the clinical behavior of BCas require further studies.



Similar content being viewed by others
Data availability
The dataset is available upon reasonable request to the corresponding author.
Change history
02 January 2022
A Correction to this paper has been published: https://doi.org/10.1007/s11033-021-07052-y
Abbreviations
- ACTRT3:
-
Actin Related Protein T3; 3q26.2
- APOBR:
-
Apolipoprotein B Receptor
- ARNT:
-
Aryl hydrocarbon receptor nuclear translocator
- BCa:
-
Bladder cancer
- BER:
-
Base excision repair
- BLNK:
-
B cell linker
- COMT:
-
Catechol-O-methyltransferase
- CYP1B1:
-
Cytochrome P450 family 1 subfamily B member 1
- CYP3A4:
-
Cytochrome P450 3A4
- dbSNP:
-
Database of single nucleotide polymorphisms
- DNA:
-
Deoxyribonucleic acid
- DRC:
-
DNA Repair Capacity
- DSBR:
-
Double-strand break repair
- EDTA:
-
Ethylene diamine tetra-acetic acid
- ERCC4:
-
Excision Repair Cross-Complementation group 4
- GSTM1:
-
Glutathione-s-transferase M1
- GWAS:
-
Genome-Wide Association Study
- GSS:
-
Glutathione Synthase
- GSTA4:
-
Glutathione S-transferase Alpha 4
- HG:
-
High-Grade
- IARC:
-
International Agency for Research on Cancer
- IL27:
-
Interleukin 27
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- LG:
-
Low-Grade
- MIBC:
-
Muscle-invasive BCa
- MMR:
-
Mismatch repair
- MPG:
-
N-methylpurine DNA glycosylase
- NEIL2:
-
Nei-like DNA glycosylase 2
- NAT2:
-
N-acetyltransferase 2
- NER:
-
Nucleotide excision repair
- NGS:
-
Next-Generation Sequencing
- NMIBC:
-
Non-muscle-invasive BCa
- PRKAR2B:
-
Protein Kinase cAMP-dependent type II regulatory subunit beta
- ROCK1:
-
Rho associated coiled-coil containing protein kinase 1
- SNP:
-
Nucleotide polymorphisms
- TCR:
-
Transcription-coupled repair
- TP63:
-
Tumor Protein p63
- VPS37C:
-
Vacuolar protein sorting-associated protein 37C
- WHO:
-
World Health Organization
- XPC:
-
Xeroderma Pigmentosum, Complementation group C
- XPF:
-
Excision repair cross-complementing rodent repair deficiency, complementation group 4
References
Antoni S, Ferlay J, Soerjomataram I et al (2017) Bladder cancer incidence and mortality: a global overview and recent trends. Eur Urol 71:96–108. https://doi.org/10.1016/j.eururo.2016.06.010
Wong MCS, Fung FDH, Leung C et al (2018) The global epidemiology of bladder cancer: a joinpoint regression analysis of its incidence and mortality trends and projection. Sci Rep. https://doi.org/10.1038/s41598-018-19199-z
Cancer today. http://gco.iarc.fr/today/home. Accessed 15 Jun 2020
Bray F, Ferlay J, Soerjomataram I et al (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J Clin 68:394–424. https://doi.org/10.3322/caac.21492
Cassell A, Yunusa B, Jalloh M et al (2019) Non-muscle invasive bladder cancer: a review of the current trend in Africa. World J Oncol 10:123–131. https://doi.org/10.14740/wjon1210
Burger M, Catto JWF, Dalbagni G et al (2013) Epidemiology and risk factors of urothelial bladder cancer. Eur Urol 63:234–241. https://doi.org/10.1016/j.eururo.2012.07.033
Skipper PL, Kim MY, Sun H-LP et al (2010) Monocyclic aromatic amines as potential human carcinogens: old is new again. Carcinogenesis 31:50–58. https://doi.org/10.1093/carcin/bgp267
Matic M, Pekmezovic T, Djukic T et al (2013) GSTA1, GSTM1, GSTP1, and GSTT1 polymorphisms and susceptibility to smoking-related bladder cancer: a case–control study. Urolo Oncol 31:1184–1192. https://doi.org/10.1016/j.urolonc.2011.08.005
García-Closas M, Malats N, Silverman D et al (2005) NAT2 slow acetylation and GSTM1 null genotypes increase bladder cancer risk: results from the Spanish bladder cancer study and meta-analyses. Lancet 366:649–659. https://doi.org/10.1016/S0140-6736(05)67137-1
Kumar N, Moreno NC, Feltes BC et al (2020) Cooperation and interplay between base and nucleotide excision repair pathways: from DNA lesions to proteins. Genet Mol Biol. https://doi.org/10.1590/1678-4685-gmb-2019-0104
Greenman C, Stephens P, Smith R et al (2007) Patterns of somatic mutation in human cancer genomes. Nature 446:153–158. https://doi.org/10.1038/nature05610
Pleasance ED, Cheetham RK, Stephens PJ et al (2010) A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463:191–196. https://doi.org/10.1038/nature08658
He J, Shi T-Y, Zhu M-L et al (2013) Associations of Lys939Gln and Ala499Val polymorphisms of the XPC gene with cancer susceptibility: a meta-analysis. Int J Cancer 133:1765–1775. https://doi.org/10.1002/ijc.28089
Rouissi K, Bahria IB, Bougatef K et al (2011) The effect of tobacco, XPC, ERCC2 and ERCC5 genetic variants in bladder cancer development. BMC Cancer 11:101. https://doi.org/10.1186/1471-2407-11-101
Leibovici D, Grossman HB, Dinney CP et al (2005) Polymorphisms in inflammation genes and bladder cancer: from initiation to recurrence, progression, and survival. JCO 23:5746–5756. https://doi.org/10.1200/JCO.2005.01.598
Kanehisa M, Sato Y, Furumichi M et al (2019) New approach for understanding genome variations in KEGG. Nucleic Acids Res 47:D590–D595. https://doi.org/10.1093/nar/gky962
PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6323939/. Accessed 30 Sep 2020
Ashburner M, Ball CA, Blake JA et al (2000) Gene ontology: tool for the unification of biology. Gene Ontol Consort Nat Genet 25:25–29. https://doi.org/10.1038/75556
Jassal B, Matthews L, Viteri G et al (2020) The reactome pathway knowledgebase. Nucleic Acids Res 48:D498–D503. https://doi.org/10.1093/nar/gkz1031
Selinski S (2017) Discovering urinary bladder cancer risk variants: status quo after almost ten years of genome-wide association studies. EXCLI J 16:1288–1296. https://doi.org/10.17179/excli2017-1000
Calvez-Kelm FL, Foll M, Wozniak MB et al (2016) KRAS mutations in blood circulating cell-free DNA: a pancreatic cancer case-control. Oncotarget 7:78827–78840. https://doi.org/10.18632/oncotarget.12386
Wang M, Wang M, Zhang W et al (2009) Common genetic variants on 8q24 contribute to susceptibility to bladder cancer in a Chinese population. Carcinogenesis 30:991–996. https://doi.org/10.1093/carcin/bgp091
Dally H, Edler L, Jäger B et al (2003) The CYP3A4*1B allele increases risk for small cell lung cancer: effect of gender and smoking dose. Pharmacogenetics 13:607–618. https://doi.org/10.1097/00008571-200310000-00004
Božina N, Bradamante V, Lovrić M (2009) Genetic polymorphism of metabolic enzymes P450 (CYP) as a susceptibility factor for drug response, toxicity, and cancer risk. Arch Ind Hyg Toxicol 60:217–242. https://doi.org/10.2478/10004-1254-60-2009-1885
Yamada S, Tang M, Richardson K et al (2009) Genetic variations of NAT2 and CYP2E1 and isoniazid hepatotoxicity in a diverse population. Pharmacogenomics 10:1433–1445. https://doi.org/10.2217/pgs.09.66
Quan L, Chattopadhyay K, Nelson HH et al (2016) Differential association for N-acetyltransferase 2 genotype and phenotype with bladder cancer risk in Chinese population. Oncotarget 7:40012–40024. https://doi.org/10.18632/oncotarget.9475
Chen Y, Yu X, Li T et al (2016) Significant association of catechol-o-methyltransferase val158met polymorphism with bladder cancer instead of prostate and kidney cancer. Int J Biol Markers 31:110–117. https://doi.org/10.5301/jbm.5000204
Chen J, Lipska BK, Halim N et al (2004) Functional analysis of genetic variation in catechol-o-methyltransferase (COMT): effects on mRNA, protein, and enzyme activity in postmortem human brain. Am J Hum Genet 75:807–821
Figueroa JD, Han SS, Garcia-Closas M et al (2014) Genome-wide interaction study of smoking and bladder cancer risk. Carcinogenesis 35:1737–1744. https://doi.org/10.1093/carcin/bgu064
ARNT Gene—GeneCards|ARNT Protein|ARNT Antibody. https://www.genecards.org/cgi-bin/carddisp.pl?gene=ARNT. Accessed 14 Feb 2021
Ke H-L, Lin J, Ye Y et al (2015) Genetic variations in glutathione pathway genes predict cancer recurrence in patients treated with transurethral resection and Bacillus Calmette-Guerin instillation for non-muscle invasive bladder cancer. Ann Surg Oncol 22:4104–4110. https://doi.org/10.1245/s10434-015-4431-5
Ateş NA, Tamer L, Ateş C et al (2005) Glutathione S-transferase M1, T1, P1 genotypes and risk for development of colorectal cancer. Biochem Genet 43:149–163. https://doi.org/10.1007/s10528-005-1508-z
Hazra TK, Kow YW, Hatahet Z et al (2002) Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J Biol Chem 277:30417–30420. https://doi.org/10.1074/jbc.C200355200
Wei H, Kamat A, Chen M et al (2012) Association of polymorphisms in oxidative stress genes with clinical outcomes for bladder cancer treated with Bacillus Calmette-Guérin. PLoS ONE. https://doi.org/10.1371/journal.pone.0038533
Kang L, Zou X, Zhang G et al (2019) A variant in a microRNA binding site in NEIL2 3′UTR confers susceptibility to age-related cataracts. FASEB J 33:10469–10476. https://doi.org/10.1096/fj.201802291R
Sugasawa K, Ng JMY, Masutani C et al (1998) Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol Cell 2:223–232. https://doi.org/10.1016/S1097-2765(00)80132-X
Zhu Y, Lai M, Yang H et al (2007) Genotypes, haplotypes and diplotypes of XPC and risk of bladder cancer. Carcinogenesis 28:698–703. https://doi.org/10.1093/carcin/bgl201
Dai Y, Song Z, Zhang J, Gao W (2019) Comprehensive assessment of the association between XPC rs2228000 and cancer susceptibility based on 26835 cancer cases and 37069 controls. Biosci Rep. https://doi.org/10.1042/BSR20192452
Zhang J-S, Zhang C, Yan X-Y et al (2013) Effect of Xeroderma pigmentosum complementation group F polymorphisms on gastric cancer risk and associations with H.pylori infection. Asian Pac J Cancer Prev 14:1847–1850. https://doi.org/10.7314/apjcp.2013.14.3.1847
Shi T-Y, He J, Qiu L-X et al (2012) Association between XPF polymorphisms and cancer risk: a meta-analysis. PLoS ONE 7:e38606. https://doi.org/10.1371/journal.pone.0038606
Association between XPF Polymorphisms and Cancer Risk: A Meta-Analysis. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3388076/. Accessed 10 Mar 2020
Xu X-P, Hua L-Y, Chao H-L et al (2017) Genetic association between IL-27 rs153109 polymorphism and cancer risk in Chinese population: a meta-analysis. J Recept Signal Trans 37:335–340. https://doi.org/10.3109/10799893.2014.986743
Masson-Lecomte A, de Maturana EL, Goddard ME et al (2016) Inflammatory-related genetic variants in non–muscle-invasive bladder cancer prognosis: a multimarker bayesian assessment. Cancer Epidemiol Biomarkers Prev 25:1144–1150. https://doi.org/10.1158/1055-9965.EPI-15-0894
Minegishi Y, Rohrer J, Coustan-Smith E et al (1999) An essential role for BLNK in human B cell development. Science 286:1954–1957. https://doi.org/10.1126/science.286.5446.1954
Nakayama J, Yamamoto M, Hayashi K et al (2009) BLNK suppresses pre–B-cell leukemogenesis through inhibition of JAK3. Blood 113:1483–1492. https://doi.org/10.1182/blood-2008-07-166355
Expression of BLNK in cancer—Summary—The Human Protein Atlas. https://www.proteinatlas.org/ENSG00000095585-BLNK/pathology. Accessed 13 Jul 2020
Bergholz J, Xiao Z-X (2012) Role of p63 in development, tumorigenesis and cancer progression. Cancer Microenviron 5:311–322. https://doi.org/10.1007/s12307-012-0116-9
Urist MJ, Di Como CJ, Lu M-L et al (2002) Loss of p63 expression Is associated with tumor progression in bladder cancer. Am J Pathol 161:1199–1206. https://doi.org/10.1016/S0002-9440(10)64396-9
Kiemeney LA, Thorlacius S, Sulem P et al (2008) Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat Genet 40:1307–1312. https://doi.org/10.1038/ng.229
Lehmann M-L, Selinski S, Blaszkewicz M et al (2010) Rs710521[A] on chromosome 3q28 close to TP63 is associated with increased urinary bladder cancer risk. Arch Toxicol 84:967–978. https://doi.org/10.1007/s00204-010-0617-6
Yang S, Zhao Y, Tian Y et al (2018) Common variants of ROCKs and the risk of hypertension, and stroke: two case-control studies and a follow-up study in Chinese Han population. Biochimica et Biophysica Acta (BBA) 1864:778–783. https://doi.org/10.1016/j.bbadis.2017.12.007
Lochhead PA, Wickman G, Mezna M, Olson MF (2010) Activating ROCK1 somatic mutations in human cancer. Oncogene 29:2591–2598. https://doi.org/10.1038/onc.2010.3
Figueroa JD, Ye Y, Siddiq A et al (2014) Genome-wide association study identifies multiple loci associated with bladder cancer risk. Hum Mol Genet 23:1387–1398. https://doi.org/10.1093/hmg/ddt519
Wang M, Chu H, Lv Q et al (2014) Cumulative effect of genome-wide association study-identified genetic variants for bladder cancer. Int J Cancer 135:2653–2660. https://doi.org/10.1002/ijc.28898
Simonis K, Shariat SF, Rink M, Urothelial Cancer Working Group of the Young Academic Urologists (YAU) Working Party of the European Association of Urology (EAU) (2014) Smoking and smoking cessation effects on oncological outcomes in nonmuscle invasive bladder cancer. Curr Opin Urol 24:492–499. https://doi.org/10.1097/MOU.0000000000000079
Talhout R, Schulz T, Florek E et al (2011) Hazardous compounds in tobacco smoke. Int J Environ Res Public Health 8:613–628. https://doi.org/10.3390/ijerph8020613
Wang L, Hu Z, Deng X et al (2013) Association between common CYP1A2 polymorphisms and theophylline metabolism in non-smoking healthy volunteers. Basic Clin Pharmacol Toxicol 112:257–263. https://doi.org/10.1111/bcpt.12038
Acknowledgements
The authors thank all patients for participating in this study. We greatly thank also the Urology department, Charles Nicolle hospital, Tunis, Tunisia, Biochemistry Department, Charles Nicolle Hospital, Tunis and the International Agency for Research on Cancer IARC; Lyon, France.
Funding
No funding was obtained for this study.
Author information
Authors and Affiliations
Contributions
IH: Collecting clinical samples and data, designed the panel, molecular analysis, drafted the manuscript and bioinformatics analyzes. SB, HD and FH: Help in Bioinformatics and statistical analyzes. GD and CV: Technical assistance for NGS analyzes. HA: Clinical characterization of the studied population. ZN and MA: Help in collecting samples. MC and JM: Designed the study and revised the manuscript. FL: Drafted and revised the manuscript. SO: Designed the study, performed the statistical analysis and drafted and revised the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This trial protocol and recruitment were approved and carried out after the agreement of the ethics committee of the Charles Nicolle Hospital and approved by an Ethics Committee (IEC Project No. 17-35) and an MTA [MATERIAL TRANSFER AGREEMENT MTA/2017/IMP/GCS)/0356] from IARC.
Consent to participate
Informed consent was obtained from all participants (in both languages: Arabic/French) prior to enrollment and participation in this study. All samples are coded and no patient names appear in the study. All data whether clinical or personal remains anonymous and secret.
Consent for publication
Not applicable. This study did not use identifying images and any clinical and personal details despite written informed consent for publication of their clinical details and/or clinical.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised due to one of the co-authors′ last names, Florence Le CalvezKelm, was misspelled.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hemissi, I., Boussetta, S., Dallali, H. et al. Development of a custom next-generation sequencing panel for the determination of bladder cancer risk in a Tunisian cohort. Mol Biol Rep 49, 1233–1258 (2022). https://doi.org/10.1007/s11033-021-06951-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06951-4