Abstract
Background
Noncatalytic region of tyrosine kinase 1 (NCK1) plays a key role in extracellular matrix degradation, which is required for the metastasis of triple-negative breast cancer (TNBC). However, the role NCK1 plays in the metastatic progression of TNBC is unknown.
Methods and results
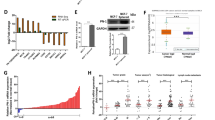
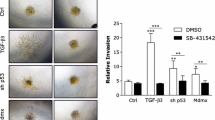
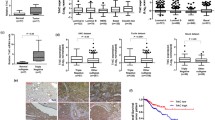
Based on online databases, NCK1 was found to be highly expressed in TNBC as compared to normal breast-like subjects, which was also confirmed using TNBC cells and a tissue microarray. NCK1 expression gradually decreased with increased tumor stage. High NCK1 expression displayed a poor prognosis in lymph node-positive metastatic TNBC patients, but not in lymph node-negative patients. Using transwell assays and immunoblotting, we confirmed that NCK1 overexpression promoted, while NCK1 downregulation inhibited migration capabilities, as well as the expression of matrix metalloproteinases (MMP2/9), uridylyl phosphate adenosine, and plasminogen activator inhibitor-1 in TNBC cells. Mechanistically, NCK1 upregulation mediated the activation of MMP2/9 through ERK1/2 activity. Signal transducer and activator of transcription 3 (STAT3) was positively correlated with NCK1. STAT3 could directly bind to the promoter region of NCK1 to promote its expression and was accompanied by the elevation of MMP2/9 and ERK1/2 signaling, which were partially abolished by the knockdown of NCK1 in TNBC cells.
Conclusions
NCK1 may serve as a diagnostic and prognostic marker of metastatic TNBC. STAT3 upregulation promoted the expression of NCK1, which subsequently induced the migration and activity of MMPs in a ERK1/2 signaling-dependent manner in TNBC cells. NCK1 is a promising target for improving TNBC migration.





Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article and its additional files.
Abbreviations
- TNBC:
-
Triple-negative breast cancer
- ECM:
-
Extracellular matrix
- NCK1:
-
Noncatalytic region of tyrosine kinase 1
- TCGA:
-
The Cancer Genome Atlas
- GEPIA:
-
Gene Expression Profiling Interactive Analysis
- MMPs:
-
Matrix metalloproteinases
- uPA:
-
Uridylyl phosphate adenosine
- PAI-1:
-
Plasminogen activator inhibitor-1
- ER:
-
Estrogen receptor
- PR:
-
Progesterone receptor
- HER2:
-
Receptor tyrosine-protein kinase erbB-2
- BRCA:
-
Breast carcinoma
- CSCC:
-
Cervical squamous cell carcinoma
- CRC:
-
Colorectal cancer
- STAT3:
-
Signal transducer and activator of transcription 3
- ERK:
-
Extracellular signal-regulated kinase
- FGF2:
-
Fibroblast growth factor 2
References
Figueroa-Magalhaes MC, Jelovac D, Connolly RM et al (2014) Treatment of HER2-positive breast cancer. Breast 23:128–136
Maumy L, Harrissart G, Dewaele P et al (2019) Impact of nutrition on breast cancer mortality and risk of recurrence, a review of the evidence. Bull Cancer 107(1):61–71
Igissinov N, Toguzbayeva A, Turdaliyeva B et al (2019) Breast cancer in megapolises of Kazakhstan: epidemiological assessment of incidence and mortality. Iran J Public Health 48:1257–1264
Deng L, Lei QQ, Wang Y et al (2017) Downregulation of miR-221-3p and upregulation of its target gene PARP1 are prognostic biomarkers for triple negative breast cancer patients and associated with poor prognosis. Oncotarget 8:108712–108725
O’Brien KM, Cole SR, Tse CK et al (2010) Intrinsic breast tumor subtypes, race, and long-term survival in the Carolina Breast Cancer Study. Clin Cancer Res 16:6100–6110
Hayashi Y, Satake H, Ishigaki S et al (2019) Kinetic volume analysis on dynamic contrast-enhanced MRI of Triple-Negative breast cancer: associations with survival outcomes. Br J Radiol 92(1106):20190712
Wang QM, Lv L, Tang Y et al (2019) MMP-1 is overexpressed in triple-negative breast cancer tissues and the knockdown of MMP-1 expression inhibits tumor cell malignant behaviors in vitro. Oncol Lett 17:1732–1740
Koblinski JE, Ahram M, Sloane BF (2000) Unraveling the role of proteases in cancer. Clin Chim Acta 291:113–135
Golubkov VS, Strongin AY (2007) Proteolysis-driven oncogenesis. Cell Cycle 6:147–150
Visse R, Nagase H (2003) Matrix metalloproteinases and tissue inhibitors of metalloproteinases: structure, function, and biochemistry. Circ Res 92:827–839
Brinckerhoff CE, Rutter JL, Benbow U (2000) Interstitial collagenases as markers of tumor progression. Clin Cancer Res 6:4823–4830
Li W, She H (2000) The SH2 and SH3 adapter Nck: a two-gene family and a linker between tyrosine kinases and multiple signaling networks. Histol Histopathol 15:947–955
Labelle-Cote M, Dusseault J, Ismail S et al (2011) Nck2 promotes human melanoma cell proliferation, migration and invasion in vitro and primary melanoma-derived tumor growth in vivo. BMC Cancer 11(1):1–19
Katayama MLH, Vieira RAC, Andrade VP et al (2019) Stromal cell signature associated with response to neoadjuvant chemotherapy in locally advanced breast cancer. Cells 8(12):1566
Oser M, Dovas A, Cox D et al (2011) Nck1 and Grb2 localization patterns can distinguish invadopodia from podosomes. Eur J Cell Biol 90:181–188
Stylli SS, Stacey TTI, Verhagen AM et al (2009) Nck adaptor proteins link Tks5 to invadopodia actin regulation and ECM degradation. J Cell Sci 122:2727–2740
Oser M, Mader CC, Gil-Henn H et al (2010) Specific tyrosine phosphorylation sites on cortactin regulate Nck1-dependent actin polymerization in invadopodia. J Cell Sci 123:3662–3673
Furtek SL, Backos DS, Matheson CJ et al (2016) Strategies and approaches of targeting STAT3 for cancer treatment. ACS Chem Biol 11:308–318
Huynh J, Chand A, Gough D et al (2019) Therapeutically exploiting STAT3 activity in cancer—using tissue repair as a road map. Nat Rev Cancer 19:82–96
Sirkisoon SR, Carpenter RL, Rimkus T et al (2018) Interaction between STAT3 and GLI1/tGLI1 oncogenic transcription factors promotes the aggressiveness of triple-negative breast cancers and HER2-enriched breast cancer. Oncogene 37:2502–2514
Kamran MZ, Patil P, and Gude RP (2013) Role of STAT3 in cancer metastasis and translational advances. Biomed Res Int
Tang ZF, Li CW, Kang BX et al (2017) GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 45:W98–W102
Saini KS, Taylor C, Ramirez AJ et al (2012) Role of the multidisciplinary team in breast cancer management: results from a large international survey involving 39 countries. Ann Oncol 23:853–859
Bhattacharjee S, Nandi S (2017) Synthetic lethality in DNA repair network: a novel avenue in targeted cancer therapy and combination therapeutics. IUBMB Life 69:929–937
Bhattacharjee S, Nandi S (2016) Choices have consequences: the nexus between DNA repair pathways and genomic instability in cancer. Clin Transl Med 5:45
Bhattacharjee S, Nandi S (2017) DNA damage response and cancer therapeutics through the lens of the Fanconi Anemia DNA repair pathway. Cell Commun Signal 15(1):1–10
Bhattacharjee S, Nandi S (2018) Rare genetic diseases with defects in DNA repair: opportunities and challenges in orphan drug development for targeted cancer therapy. Cancers 10(9):298
Xia P, Huang MC, Zhang YT et al (2019) NCK1 promotes the angiogenesis of cervical squamous carcinoma via Rac1/PAK1/MMP2 signal pathway. Gynecol Oncol 152:387–395
Liu XH, Zhang J, Duan ZN et al (2020) Nck1 promotes the progression of ovarian carcinoma by enhancing the PI3K/AKT/p70S6K signaling. Hum Cell 33:768–779
Yin J, Wu K, Ma QY et al (2019) Revisiting non-BRCA1/2 familial whole exome sequencing datasets implicates NCK1 as a cancer gene. Front Genet 10:527
West NW, Garcia-Vargas A, Chalfant CE et al (2013) OSU-03012 sensitizes breast cancers to lapatinib-induced cell killing: a role for Nck1 but not Nck2. BMC Cancer 13:256
Deshpande RP, Panigrahi M, Sekhar YBVKC et al (2019) Expression and clinicopathological significance of Nck1 in human astrocytoma progression. Int J Neurosci 129:171–178
Fallahi H, Godini R (2019) System-level responses to cisplatin in pro-apoptotic stages of breast cancer MCF-7 cell line. Comput Biol Chem 83:107155
Ruusala A, Pawson T, Heldin CH et al (2008) Nck adapters are involved in the formation of dorsal ruffles, cell migration, and Rho signaling downstream of the platelet-derived growth factor beta receptor. J Biol Chem 283:30034–30044
Zhou Q, Huang T, Wang YF et al (2012) Identification of Max binding protein as a novel binding protein of Nck1 and characterization of its role in inhibiting human liver cancer SK-HEP-1 cells. Chin Med J 125:3336–3339
Li H, Li B, Larose L (2017) IRE1 alpha links Nck1 deficiency to attenuated PTP1B expression in HepG2 cells. Cell Signal 36:79–90
Miyamoto Y, Torii T, Kawahara K et al (2016) Dock8 interacts with Nck1 in mediating Schwann cell precursor migration. Biochem Biophys Rep 6:113–123
Huang M, Anand S, Murphy EA et al (2012) EGFR-dependent pancreatic carcinoma cell metastasis through Rap1 activation. Oncogene 31:2783–2793
Morris DC, Popp JL, Tang LK et al (2017) Nck deficiency is associated with delayed breast carcinoma progression and reduced metastasis. Mol Biol Cell 28:3500–3516
Chaki SP, Barhoumi R, Rivera GM (2019) Nck adapter proteins promote podosome biogenesis facilitating extracellular matrix degradation and cancer invasion. Cancer Med 8:7385–7398
Linder S (2009) Invadosomes at a glance. J Cell Sci 122:3009–3013
Yan GR, Yin XF, Xiao CL et al (2011) Identification of novel signaling components in genistein-regulated signaling pathways by quantitative phosphoproteomics. J Proteomics 75:695–707
Anselmi F, Orlandini M, Rocchigiani M et al (2012) c-ABL modulates MAP kinases activation downstream of VEGFR-2 signaling by direct phosphorylation of the adaptor proteins GRB2 and NCK1. Angiogenesis 15:187–197
Yang CC, Yu HC, Chen R et al (2019) CXCL1 stimulates migration and invasion in ER-negative breast cancer cells via activation of the ERK/MMP2/9 signaling axis. Int J Oncol 55:684–696
Li B, Pi ZJ, Liu L et al (2013) FGF-2 prevents cancer cells from ER stress-mediated apoptosis via enhancing proteasome-mediated Nck degradation. Biochem J 452:139–145
Sola-Penna M, Paixao LP, Branco JR et al (2020) Serotonin activates glycolysis and mitochondria biogenesis in human breast cancer cells through activation of the Jak1/STAT3/ERK1/2 and adenylate cyclase/PKA, respectively. Br J Cancer 122:194–208
Lv JX, Yu W, Zhang YA et al (2020) LNK promotes the growth and metastasis of triple negative breast cancer via activating JAK/STAT3 and ERK1/2 pathway. Cancer Cell Int 20(1):1–13
Zhang F, Lu YX, Chen Q et al (2017) Identification of NCK1 as a novel downstream effector of STAT3 in colorectal cancer metastasis and angiogenesis. Cell Signal 36:67–78
Acknowledgements
We thank the pathologists Chi-Meng Tzeng, Zhiming Zhang, Xianyang Luo and Shuai Chen for analyzing the Immunohistochemical results. We also thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Funding
This work was supported by Youth Fund of Pingdingshan University (PXY-QNJJ-202108) and Henan Provincial Key Science and Technology Research Projects (No. 192102310087).
Author information
Authors and Affiliations
Contributions
ZXH conceived the idea and designed the project; HPN performed the in vitro experiments and analysed the data based on the online database; SJY and QJX performed the Transwell assay; BXG performed the qRT-PCR assay; LJX determined the cell viability measurement. WFB and YYM collected the online data and performed the IHC staining assay. ZXH and HPN drafted the text; All authors edited and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All authors have completed the ICMJE uniform disclosure form. The authors declare that they have no competing interests.
Ethical approval
All experiments were approved by the Ethics Committee of Pingdingshan University.
Informed consent
The data analyzed in this study all were downloaded from online database. And the study did implicate in clinical trials. All authors are in agreement with the content of the manuscript. All authors discussed the results and edited this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11033_2021_6868_MOESM1_ESM.tif
Supplementary file1 Figure S1. Clinical value of NCK1 in breast cancer subjects. (A) TNBC patients were divided into three classifications based on different tumor stages (according to SBR classified method: SBR1 (n=544), SBR2 (n=1699), and SBR3 (n=1374)). Subsequently, multiple comparisons between NCK1 expression and SBR classifications were determined by an one-way ANOVA using Tukey’s test. (B) Survival analysis was determined using GEPIA 2.0 in breast cancer patients with low (n=534) and high NCK1 expression (n=534). (TIF 424 kb)
11033_2021_6868_MOESM2_ESM.tif
Supplementary file2 Supplementary Figure 2 Effect of NCK1 on cell proliferation of TNBC cells. MDA-MB-468 and BT549 cells were transfected with NCK1-overexpressed plasmid or NCK1 shRNA. Then cell viabilities in MDA-MB-468 (A) and BT549 (B) cells were determined by CCK8 assay. All assays were performed three times (n=3). (TIF 363 kb)
11033_2021_6868_MOESM3_ESM.tif
Supplementary file3 Figure S3. Effect of NCK1 on the cell invasion of TNBC cells. MDA-MB-468 cells were transfected with NCK1-overexpressing plasmid or NCK1 shRNA. (A) Then the cell invasion in MDA-MB-468 cells was determined by a transwell assay. All assays were performed three times (n=3). (B) The relative quantification of the invasiveness of the cells is shown on the right. Six random sights of each group were selected to conduct the semi-quantitative analysis. **P < 0.01. (TIF 1833 kb)
11033_2021_6868_MOESM4_ESM.tif
Supplementary file4 Figure S4. Association of STAT3 and NCK1 in TNBC and non-TNBC cells. (A) BT-474 cells were transfected with empty vectors, STAT3-overexpressing plasmid, or NCK1 sRNA, and then 48 h later, the luciferase activity of NCK1 was determined by the Dual Luciferase Reporter Assay Kit. All assays were performed three times (n=3). (B) BT-474 cells were treated with or without IL-6 for 30 min. The direct binding of STAT3 to NCK1, the direct binding of STAT3 to SOCS3, and the direct binding of STAT3 to α satellite in BT-474 cells were measured by a chromatin immunoprecipitation (ChIP) assay. All assays were performed three times (n=3). (C) BT-549 cells were transfected with empty vectors, STAT3-overexpressing plasmid, or NCK1 sRNA. Subsequently, protein expression of STAT3, ERK1/2, phosphorylated ERK1/2, NCK1, MMP-9, and MMP-2 was measured by WB. All assays were performed three times (n=3). The relative quantitative analysis of phosphorylated ERK1/2, STAT3, MMP9/2, and NCK1 is shown in panel C. (D) BT-474 cells were transfected with empty vectors, STAT3-overexpressing plasmid, or NCK1 sRNA. Subsequently, the protein expression of STAT3, ERK1/2, phosphorylated ERK1/2, NCK1, MMP-9, and MMP-2 was measured by WB. All assays were performed three times (n=3). The relative quantitative analysis of phosphorylated ERK1/2, STAT3, MMP9/2, and NCK1 is shown in panel D. *P < 0.05; **P < 0.01. (TIF 1110 kb)
Rights and permissions
About this article
Cite this article
He, P., Sheng, J., Qi, J. et al. STAT3-induced NCK1 elevation promotes migration of triple-negative breast cancer cells via regulating ERK1/2 signaling. Mol Biol Rep 49, 267–278 (2022). https://doi.org/10.1007/s11033-021-06868-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06868-y