Abstract
Background
Meloidogyne arenaria is an economically important root-knot nematode (RKN) species whose hosts include maize (Zea mays). The plant response to RKN infection activates many cellular mechanisms, among others, changes in the expression level of genes encoding transcription and elongation factors as well as proteins related to cell wall organization.
Methods and results
This study is aimed at characterization of expression of selected transcription and elongation factors encoding the genes WRKY53, EF1a, and EF1b as well as the ones encoding two proteins associated with cell wall functioning (glycine-rich RNA-binding protein, GRP and polygalacturonase, PG) during the maize response to M. arenaria infection. The changes in the relative level of expression of genes encoding these proteins were assessed using the reverse transcription-quantitative real-time PCR. The material studied were leaves and root samples collected from four maize varieties showing different susceptibilities toward M. arenaria infection, harvested at three different time points. Significant changes in the expression level of GRP between susceptible and tolerant varieties were observed.
Conclusions
Results obtained in the study suggest pronounced involvement of glycine-rich RNA-binding protein and EF1b in the maize response and resistance to RKN.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Root-knot nematodes (RKNs) include more than 60 described Meloidogyne (Tylenchidae: Tylenchus) species [1]. These nematodes are highly polyphagous with a very wide host range, comprising both, mono- and dicotyledonous plants [2]. Meloidogyne arenaria, along with M. incognita, M. javanica, and M. hapla, is one of the most economically important RKN species [1]. This species is distributed worldwide on most continents and parasitizes many major food crops and ornamental plants grown in tropical, subtropical, and temperate climates, in fields as well as in greenhouses. One of the main monocotyledonous hosts of M. arenaria is maize (Zea mays) [3], which is also one of the most important food crops worldwide in human and animal nutrition.
Meloidogyne arenaria is invasive in the J2 larvae stadium. To get into the root tissue, RKNs use their hollow mouth stylet and after mechanical penetration inject the cell wall-degrading enzymes, effectors, and other virulence factors into the cell [1]. During the invasion, this nematode induces changes in expression of a broad spectrum of genes engaged in numerous processes, including in wound and defense responses, reorganization of the cell wall, cell cycle and cytoskeleton organization to establish giant cells, which in consequence leads to galls formation [4]. The presence of nematodes is recognized by the plant through the perception of the pathogen-associated molecular patterns (PAMPs) located on their surfaces. After PAMPs recognition by plant cells, the first layer of defense response is induced [5].
Many aspects of plant response to nematode infection are still not fully understood. Analyses are conducted on monocotyledonous as well as on dicotyledonous hosts and some similarities and differences in the response between these two groups have been observed [6]. For instance, some studies conducted on monocotyledonous plants indicated activation of jasmonic acid (JA)- and salicylic acid (SA)- mediated pathways in RKN-plant interactions. In compatible interactions, the expression level of genes encoding proteins associated with these pathways was upregulated only in the early stages of infection and found to be suppressed later on, in contrast to incompatible interactions in which upregulation of these genes was observed in later stages as well [7,8,9].
The plant response to RKNs is known to involve many proteins, including a number of transcription factors that have been established to play important roles in plant-nematode interactions. Interestingly, some differences between mono- and dicotyledonous hosts' response to nematode infection concerning changes in the expression of genes encoding these proteins have also been observed [10, 11]. One of the frequently analyzed groups of transcription factors (TFs) are those that are crucial in the regulation of gene expression and are often described as down-stream defense-responsive genes engaged in the response to biotic and abiotic stress conditions [12]. WRKY family is a major group of transcription factors that may act as positive or negative regulators of both components of the plant innate immunity: pathogen-associated molecular pattern-triggered immunity (PTI) as well as in effector-triggered immunity (ETI) [13]. The involvement of WRKYs has been also reported in development of the host resistance to RKNs infection [14]. One of the central factors in the WRKY network, regulating among others early senescence in plants, is WRKY53 [15]. This factor has been reported also to take part downstream of salicylic acid and to be negatively regulated by signaling through jasmonic acid and ethylene in Arabidopsis [16]. Moreover, WRKY53 is induced in rice by chitin oligosaccharides and stimulates the expression of PR proteins and peroxidases [17]. However, its role in plant response to Meloidogyne infection has not been analyzed as yet.
From among TFs, the elongation factor 1 (EF1) has been established to play an important role in many processes in plants. EF1, composed of a G-protein (EF1a) and the guanine–nucleotide exchange factor (EF1b), is involved in many processes in plants, mainly in the regulation, proliferation, and differentiation of cells [18, 19]. EF1a is a multifunctional protein, which catalyzes the binding of aminoacyl tRNA to the acceptor site on the ribosome and is involved in various other cellular processes such as signal transduction or nuclear export of proteins [20, 21]. Moreover, it is a major cytoskeleton-associated protein, binding microtubules and microfilaments showing actin-binding activity [22]. It has been reported that EF1a interacts with the viral RNA-dependent RNA polymerase and the 30-terminal genomic RNA of tobacco mosaic virus (TMV) during Nicotiana benthamiana infection [23]. Interestingly, knockdown of expression of the gene encoding EF1a inhibits the cell death response in soybean and alters this host response to soybean mosaic virus [24]. On the other hand, EF1b can disrupt EF1a-induced actin organization [25]. EF1b is also engaged in the growth and cell cycle regulation [26]. However, the roles of EF1a and EF1b’s in plant-RKN interactions have not been described yet.
The frontline of the plant defense system constitutes the cell wall, whose dry mass in 90% is composed of cellulose, hemicelluloses, and pectins [27]. Polygalacturonases (PGs) are known to be engaged in the last step of pectin degradation but their role in plant development has been also described [28]. Moreover, in maize, PGs have been reported to take part in the suppression of programmed cell death [29]. There are also some data indicating the importance of plant PG during Glycine max response to Heterodera glycines infection. It has been also shown that the upregulation of PG transcription in soybean roots in the early stage of nematode infection could facilitate successful parasitism [30]. However, most of the studies on plant-pathogen interactions have been focused on the role of pathogen’s PGs and plant polygalacturonase-inhibiting proteins (PGIPs) (e.g. [31]), while the function of plant’s PGs in defense response has not been very widely analyzed. On the other hand, the role of glycine-rich proteins (GRPs) was relatively well studied in the infection process. Among others, GRPs have been suggested to initiate the recognition of the stimuli from the environment and to participate in signal transduction [32]. GRPs have been also identified as part of the defense and repair system of the plants but their mode of action on the molecular level is still not clear [33]. Moreover, the large spectrum of subcellular locations and the broad structural diversity of GRPs suggests that they are involved in several independent physiological processes [34]. Besides the functions related to cell wall functioning and plant defense response, GRPs have been described to act as extracellular ligands of kinase proteins, RNA-binding proteins during osmotic stress, and many others [35]. The regulation of RNA metabolism by glycine-rich RNA-binding protein has been also reported as important for plant innate immunity [36]. The glycine-rich RNA-binding proteins are known to be involved in post-transcriptional regulation of gene expression as well as in RNA processing, which is part of developmental regulation in plants [37].
The study presented is aimed at characterization of gene expression of the following transcription and elongation factors: WRKY53, EF1a, and EF1b and the genes encoding two proteins related to cell wall functioning: glycine-rich RNA-binding protein 2 (GRP2) and polygalacturonase (PG), during the maize response to M. arenaria infection. For the study the transcription and elongation factors genes that have not been thoroughly analyzed yet, have been chosen. The relative changes in the expression level of the genes encoding these proteins were assessed using reverse transcription-quantitative real-time PCR (RT-qPCR) using RNA isolated from leaves and root samples derived from maize varieties showing different susceptibilities toward M. arenaria infection.
Materials and methods
Material
The experiments were conducted on maize plants from four varieties of different susceptibilities toward M. arenaria infection. Two sensitive varieties: PR38F58 (Pioneer) and Tasty Sweet (Seminis) as well as two tolerant varieties: PR39A98 (Pioneer) and Multitop (Syngenta) were selected during our previous study [7].
Meloidogyne arenaria larvae were collected from tobacco roots using NaOCl, according to the technique described by Hussey and Barker [38].
Growing conditions and sample collection
The 3–4 week old seedlings of maize plants at the 4–5 leaves stage were inoculated with 1500 J2 stage larvae of M. arenaria suspended in water. From each variety, four infected and four non-infected control plants were grown at constant day/night temperatures of 25 °C/20 °C and under controlled light conditions. Samples from roots and leaves from healthy and nematode-inoculated plants were collected at three time points: 24 h (hpi), 3 days (dpi), and 7 days (dpi) after inoculation.
RNA extraction and cDNA synthesis
From all leaves and roots samples, total RNA was extracted using Spectrum™ Plant Total RNA Kit (Sigma-Aldrich). RNA concentration and purity were determined with a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). From each sample, 200 ng was used as a template for cDNA synthesis with a Maxima First Strand cDNA Synthesis Kit for RT-qPCR with dsDNase (Thermo Fisher Scientific). The cDNA has been diluted 1:1 with sterile distilled water and used as a template for the real-time PCR assay.
Real-time PCR reactions
Real-time PCR reactions were carried out with primers amplifying the reference genes: Leunig and FPGS [39] as well as genes encoding transcription and elongation factors WRKY53, EF1a, and EF1b and the genes encoding proteins connected with plant cell wall: polygalacturonase (PG) and glycine-rich RNA-binding protein 2 (GRP2), using specific primer pairs listed in the table below (Table 1).
All subsequent qPCR reactions were conducted in three biological replicates and three technical replicates, in 10 µl of reaction mixture containing: 1 μl of template cDNA, 0.5 μM of each primer, 5 μl of iTaq master mix (Biorad), and sterile distilled water. Moreover, a sample with no cDNA template was used to exclude the reagents contamination. Reactions were performed using LightCycler 96 (Roche) with the following thermal profile: 5 min at 95 °C; 40 cycles at 95 °C for 10 s, annealing temperature appropriate for each primers pair (Table 1) for 10 s and 72 °C for 10 s. The melting phase began at 65 °C and ended at 95 °C, with an increase of 1 °C at each step. Relative quantification analyses were performed using the GenEx 6.0 software (MultiD Analyses AB) using the formula: Relative quantities = 2−ΔΔCq. Expression data were normalized using the genes encoding Leunig and FPGS as references [36]. Statistical significance of down- or up-regulation was also calculated with GenEx using the t-student test and P < 0.05.
Results
The analysis presented in this study concerned the expression of genes of maize proteins from two groups: those encoding transcription and elongation factors (WRKY53, EF1a, and EF1b) and those related to plant cell wall (PG and GRP2), in the varieties of maize showing different susceptibilities to M. arenaria, at different time points after nematode infection.
Changes in the expression level of genes encoding transcription and elongation factors
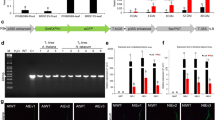
The analysis of the expression of genes encoding transcription and elongation factors revealed some changes between the samples taken at different time points after nematode inoculation as well between inoculated varieties. These differences were particularly pronounced in the level of expression of the gene encoding EF1b protein. In the samples collected from the roots, the lowest expression level was found 3 days after the infection, especially for one of the tolerant varieties—PR39A98. On the other hand, in the samples from the leaves collected from the tolerant varieties, a significant upregulation of EF1b gene expression was observed 3 dpi and was suppressed later 7 dpi. In the expression level of the gene encoding WRKY53, there were some changes between the samples collected at different time points but no significant changes between the sensitive and tolerant varieties nor between the samples from the roots and leaves were observed (Fig. 1).
Changes in the expression level of genes encoding cell wall-related proteins
Significant changes were found in the GRP2 gene expression between the samples collected from the roots and leaves as well as those taken at different time points. For the samples from the roots the changes in the level of this gene expression appeared mainly between the samples collected at different time points. In most varieties, in the first stage of infection (24 hpi), the upregulation of GRP expression occured, while downregulation was observed only in the sensitive variety—PR39F58. In the later stage of infection, 7 dpi, the expression level decreased in all varieties except the tolerant one–Multitop. In this variety, the upregulation was observed in the samples collected at all time points. In the leaves a significant difference was noted between the results obtained for the sensitive and tolerant varieties. In both tolerant varieties, PR39A98 and Multitop, the upregulation of the GRP gene expression was observed at all time points tested, in contrast to sensitive varieties. For the latter, the upregulation was observed only at the very beginning of infection (24 hpi) for PR39F58 and then the gene expression was downregulated. In the second sensitive variety, Tasty Sweet, the gene expression was suppressed at all time points. On the other hand, significant upregulation of PG gene expression was observed for one sensitive variety (PR39F58) in the early stage of infection (Fig. 2).
Discussion
Root-knot nematode infection causes numerous changes in many metabolic pathways in the host plant. Molecular mechanisms of plant-Meloidogyne interactions are still not fully understood. Many studies indicate an important role of transcription factors and cell wall-related proteins in the infection process and plant resistance to infection in dicotyledonous (e.g. [10, 40,41,42,43]) as well as in monocotyledonous hosts [8, 9, 11, 44].
In this study, we reported the upregulation of the expression level of the gene encoding WRKY53 transcription factor in one tolerant variety (Multitop) in the early stage of infection. This finding partially coincides with the results obtained for WRKY13 and WRKY24 transcription factors during interactions of RKNs and their monocotyledonous host—rice, in which the upregulation of the genes encoding these proteins was observed only in tolerant varieties in all stages of infection [8, 9, 11]. The WRKY53 protein analyzed in this study has been reported to be involved in plant response to pathogens, including bacteria [45], viruses [46], and herbivores [47] but there have been no data on its role in plant-RKNs interaction yet.
Previous analyses on tomato-M. incognita pathosystem have shown the downregulation of another gene—EF1—in the later stages of nematode infection in a susceptible variety of Solanum lycopersicum [40]. However, no data on the role of EF1a and EF1b proteins in the response of monocotyledonous to RKNs infection have been available. Our results indicated some significant changes in the expression level of gene encoding EF1b, which may suggest a role of this protein in the plant response and plant resistance to infection with M. arenaria species. In contrast to our results for biotic stress conditions, EF1a and EF1b have been described as stable genes in plants under abiotic stresses and are widely used as reference genes in qPCR reactions [48].
Two other proteins analyzed in this study are related to cell wall functioning. In our experiment the upregulation of the PG gene in the sensitive variety was observed, which corresponds with the data reported for H. glycines–G. max pathosystem, for which the upregulation of the gene encoding polygalacturonase was also observed during compatible interactions [30]. Another gene analyzed in our study was that encoding a protein belonging to GRPs family. These proteins are known to be scaffold or agglutinating agents for the deposition of cell wall constituents in plant's cell wall structure [35]. This proteins group is characterized by the presence of semi-repetitive glycine-rich motifs [34]. The role of GRPs in the response of many plant species to biotic and abiotic stress conditions has been described previously [32]. The downregulation of GRPs expression has also been observed during virus infection of tobacco [49]. Moreover, one of the glycine-rich RNA-binding proteins (AtGRP7) has been described to play either a positive or negative role in defense against different pathogens. AtGRP7 confers plant defense against a tobacco mosaic virus in tobacco as well as Pectobacterium carotovorum in Arabidopsis, but on the other hand, the same protein plays a negative role in defense against the fungus Botrytis cincera in Arabidopsis [50]. Additionally, the plant species displaying increased levels of GRP transcripts have been found more resistant to biotic and abiotic stress conditions than the wild-type plants [51, 52]. This observation is in agreement with the results obtained in our studies. During maize infection by M. arenaria we observed a significant upregulation of expression of the gene encoding a glycine-rich RNA-binding protein in the tolerant varieties at all time points. In our previous study of the same pathosystem, varieties, and time points we analyzed marker genes from JA- and SA-mediated pathways, e.g. PR1, PR3, PR4, PR5, or LOX. We observed a decrease in their expression level 24 hpi, followed by an increase in expression to the basal level 3 dpi for most of the analyzed genes associated with both, JA- and SA- pathways. Moreover, the downregulation of PR-3 and PR-4 at 24 hpi was more pronounced in the tolerant varieties than in the sensitive ones [7]. Another study of a rice-M. graminicola pathosystem has shown that JA- and SA-mediated pathways are activated during the early stage of infection in both susceptible and resistant hosts, but the responses are suppressed in later stages of infection in the susceptible varieties [8, 9]. According to these results, there is no direct correlation between the changes in GRP expression level and the expression level of marker genes related with JA- and SA- mediated pathways, because for GRP no suppression was observed and upregulation occurred at all analyzed time points.
Conclusions
The results of our study indicated a potential role of transcription and elongation factors in maize response and resistance to M. arenaria infection. However, the crucial finding of our study is that the most significant differences between susceptible and tolerant varieties were observed in the expression level of the gene encoding the glycine-rich RNA-binding protein, which was substantially higher in the tolerant varieties at all time points tested. This result is in agreement with the other studies on the GRPs role in plant response to biotic and abiotic stress conditions.
References
Jones MG, Goto DB (2011) Root-knot nematodes and giant cells. Genomics and molecular genetics of Plant-nematode Interactions. Springer, Dordrecht, pp 83–100
Moens M, Perry RN, Starr JL (2009) Meloidogyne species–a diverse group of novel and important plant parasites. Root-knot nematodes 1:483
CABI (2020) Invasive species compendium—Meloidogyne arenaria. CABI, Wallingford
Gheysen G, Fenoll C (2002) Gene expression in nematode feeding sites. Annu Rev Phytopathol 40:191–219
Manosalva P, Manohar M, Von Reuss SH, Chen S, Koch A, Kaplan F et al (2015) Conserved nematode signalling molecules elicit plant defenses and pathogen resistance. Nat Commun 6:7795
Przybylska A, Obrępalska-Stęplowska A (2020) Plant defense responses in monocotyledonous and dicotyledonous host plants during root-knot nematode infection. Plant Soil 451:239–260
Przybylska A, Kornobis F, Obrępalska-Stęplowska A (2018) Analysis of defense gene expression changes in susceptible and tolerant cultivars of maize (Zea mays) upon Meloidogyne arenaria infection. Physiol Mol Plant Pathol 103:78–83
Kumari C, Dutta TK, Gahoi S, Rao U (2017) An insight into the expression profile of defence-related genes in compatible and incompatible Oryza sativa-Meloidogyne graminicola interaction. Indian J Genet Plant Breed 77(1):42–50
Kumari C, Dutta TK, Banakar P, Rao U (2016) Comparing the defence-related gene expression changes upon root-knot nematode attack in susceptible versus resistant cultivars of rice. Sci Rep 6(1):1–13
Jammes F, Lecomte P, Almeida-Engler J, Bitton F, Martin-Magniette ML, Renou JP, Abad P, Favery B (2005) Genome-wide expression profiling of the host response to root-knot nematode infection in Arabidopsis. Plant J 44:447–458
Nguyễn PV, Bellafiore S, Petitot A-S, Haidar R, Bak A, Abed A, Gantet P, Mezzalira I, de Almeida EJ, Fernandez D (2014) Meloidogyne incognita-rice (Oryza sativa) interaction: a new model system to study plant-root-knot nematode interactions in monocotyledons. Rice 7:1–13
Xing X, Li X, Zhang M, Wang Y, Liu B, Xi Q, Zhao K, Wu Y, Yang T (2017) Transcriptome analysis of resistant and susceptible tobacco (Nicotiana tabacum) in response to root-knot nematode Meloidogyne incognita infection. Biochem Biophys Res Commun 482(4):1114–1121
Negi N, Khurana P (2021) A salicylic acid inducible mulberry WRKY transcription factor, Mi WRKY53 is involved in plant defence response. Plant Cell Rep. https://doi.org/10.1007/s00299-021-02710-8
Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15:247–258
Zentgraf U, Doll J (2019) Arabidopsis WRKY53, a node of multi-layer regulation in the network of senescence. Plants 8(12):578
Murray SL, Ingle RA, Petersen LN, Denby KJ (2007) Basal resistance against Pseudomonas syringae in Arabidopsis involves WRKY53 and a protein with homology to a nematode resistance protein. Mol Plant Microbe Interact 20(11):1431–1438
Chujo T, Takai R, Akimoto-Tomiyama C, Ando S, Minami E, Nagamura Y, Kaku H, Shibuya N, Yasuda M, Nakashita H, Umemura K, Okada A, Okada K, Nojiri H, Yamane H (2007) Involvement of the elicitor-induced gene OsWRKY53 in the expression of defense-related genes in rice. Biochim Biophys Acta Gene Struct Expr 1769(7–8):497–505
Gao Y, Ma J, Zheng JC, Chen J, Chen M, Zhou YB, Fu JD, Xu ZS, Ma YZ (2019) The elongation factor GmEF4 is involved in the response to drought and salt tolerance in soybean. Int J Mol Sci 20(12):3001
Le Sourd F, Boulben S, Le Bouffant R, Cormier P, Morales J, Belle R, Mulner-Lorillon O (2006) eEF1B: at the dawn of the 21st century. Biochim Biophys Acta Gene Struct Expr 1759(1–2):13–31
Berberich T, Sugawara K, Harada M, Kusano T (1995) Molecular cloning, characterization and expression of an elongation factor 1α gene in maize. Plant Mol Biol 29(3):611–615
Suhandono S, Apriyanto A, Ihsani N (2014) Isolation and characterization of three cassava elongation factor 1 alpha (MeEF1A) promoters. PLoS ONE 9(1):e84692
Gungabissoon RA, Khan S, Hussey PJ, Maciver SK (2001) Interaction of elongation factor 1α from Zea mays (ZmEF-1α) with F-actin and interplay with the maize actin severing protein, ZmADF3. Cell Motil Cytoskelet 49(2):104–111
Yamaji Y, Sakurai K, Hamada K, Komatsu K, Ozeki J, Yoshida A, Yoshii A, Shimizu T, Namba S, Hibi T (2010) Significance of eukaryotic translation elongation factor 1A in tobacco mosaic virus infection. Arch Virol 155(2):263–268
Luan H, Shine MB, Cui X, Chen X, Ma N, Kachroo P, Zhi H, Kachroo A (2016) The potyviral P3 protein targets eukaryotic elongation factor 1A to promote the unfolded protein response and viral pathogenesis. Plant Physiol 172(1):221–234
Pittman YR, Kandl K, Lewis M, Valente L, Kinzy TG (2009) Coordination of eukaryotic translation elongation factor 1A (eEF1A) function in actin organization and translation elongation by the guanine nucleotide exchange factor eEF1Bα. J Biol Chem 284(7):4739–4747
Hossain Z, Amyot L, McGarvey B, Gruber M, Jung J, Hannoufa A (2012) The translation elongation factor eEF-1Bβ1 is involved in cell wall biosynthesis and plant development in Arabidopsis thaliana. PLoS ONE 7(1):e30425
Malinovsky FG, Fangel JU, Willats WG (2014) The role of the cell wall in plant immunity. Front Plant Sci 5:178
Yang Y, Yu Y, Liang Y, Anderson CT, Cao J (2018) A profusion of molecular scissors for pectins: classification, expression, and functions of plant polygalacturonases. Front Plant Sci 9:1208
He Y, Karre S, Johal GS, Christensen SA, Balint-Kurti P (2019) A maize polygalacturonase functions as a suppressor of programmed cell death in plants. BMC Plant Biol 19(1):1–10
Mahalingam R, Wang G, Knap HT (1999) Polygalacturonase and polygalacturonase inhibitor protein: gene isolation and transcription in Glycine max-Heterodera glycines interactions. Mol Plant Microbe Interact 12(6):490–498
Haeger W, Henning J, Heckel DG, Pauchet Y, Kirsch R (2020) Direct evidence for a new mode of plant defense against insects via a novel polygalacturonase-inhibiting protein expression strategy. J Biol Chem 295(33):11833–11844
Czolpinska M, Rurek M (2018) Plant glycine-rich proteins in stress response: an emerging, still prospective story. Front Plant Sci 9:302
Mousavi A, Hotta Y (2005) Glycine-rich proteins. Appl Biochem Biotechnol 120:169–174
Sachetto-Martins G, Franco LO, de Oliveira DE (2000) Plant glycine-rich proteins: a family or just proteins with a common motif? Biochim Biophys Acta Gene Struct Expr 1492:1–14
Mangeon A, Junqueira RM, Sachetto-Martins G (2010) Functional diversity of the plant glycine-rich proteins superfamily. Plant Signal Behav 5:99–104
Rashid A (2016) Defense responses of plant cell wall non-catalytic proteins against pathogens. Physiol Mol Plant Pathol 94:38–46
Sanan-Mishra N, Tuteja N, Sopory SK (2002) Salinity-and ABA-induced up-regulation and light-mediated modulation of mRNA encoding glycine-rich RNA-binding protein from Sorghum bicolor. Biochem Biophys Res Commun 296(5):1063–1068
Hussey RS, Barker R (1973) A comparison of methods of collecting inocula of Meloidogyne spp., including a new technique. Plant Dis Rep 57:1025–1028
Manoli A, Sturaro A, Trevisan S, Quaggiotti S, Nonis A (2012) Evaluation of candidate reference genes for qPCR in maize. J Plant Physiol 169:807–815
Shukla N, Yadav R, Kaur P, Rasmussen S, Goel S, Agarwal M, Jagannath A, Gupta R, Kumar A (2018) Transcriptome analysis of root-knot nematode (Meloidogyne incognita)-infected tomato (Solanum lycopersicum) roots reveals complex gene expression profiles and metabolic networks of both host and nematode during susceptible and resistance responses. Mol Plant Pathol 19:615–633
Bhattarai KK, Xie Q-G, Mantelin S, Bishnoi U, Girke T, Navarre DA, Kaloshian I (2008) Tomato susceptibility to root-knot nematodes requires an intact jasmonic acid signaling pathway. Mol Plant-Microbe Interact 21:1205–1214
Postnikova OA, Hult M, Shao J, Skantar A, Nemchinov LG (2015) Transcriptome analysis of resistant and susceptible alfalfa cultivars infected with root-knot nematode Meloidogyne incognita. PLoS ONE 10:e0118269
Bar-Or C, Kapulnik Y, Koltai H (2005) A broad characterization of the transcriptional profile of the compatible tomato response to the plant parasitic root knot nematode Meloidogyne javanica. Eur J Plant Pathol 111(2):181
Castañeda NEN, Alves GSC, Almeida RM, Amorim EP, Fortes Ferreira C, Togawa RC et al (2017) Gene expression analysis in Musa acuminata during compatible interactions with Meloidogyne incognita. Ann Bot 119(5):915–930
Hu Y, Dong Q, Yu D (2012) Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci 185:288–297
Van Eck L, Schultz T, Leach JE, Scofield SR, Peairs FB, Botha AM, Lapitan NL (2010) Virus-induced gene silencing of WRKY53 and an inducible phenylalanine ammonia-lyase in wheat reduces aphid resistance. Plant Biotechnol J 8:1023–1032
Hu L, Ye M, Li R, Zhang T, Zhou G, Wang Q, Lu J, Lou Y (2015) The rice transcription factor WRKY53 suppresses herbivore-induced defenses by acting as a negative feedback modulator of mitogen-activated protein kinase activity. Plant Physiol 169:2907–2921
Linardić M, Braybrook SA (2021) Identification and selection of optimal reference genes for qPCR-based gene expression analysis in Fucus distichus under various abiotic stresses. PLoS ONE 16(4):e0233249
Obrępalska-Stęplowska A, Zmienko A, Wrzesińska B, Goralski M, Figlerowicz M, Zyprych-Walczak J, Siatkowski I, Pospieszny H (2018) The defense response of Nicotiana benthamiana to peanut stunt virus infection in the presence of symptom exacerbating satellite RNA. Viruses 10(9):449
Lee HJ, Kim JS, Yoo SJ, Kang EY, Han SH, Yang K-Y, Kim YC, Gardener BM, Kang H (2012) Different roles of glycine-rich RNA-binding protein7 in plant defense against Pectobacterium carotovorum, Botrytis cinerea, and tobacco mosaic viruses. Plant Physiol Biochem 60:46–52
Kwak KJ, Kim H-S, Jang HY, Kang H, Ahn S-J (2016) Diverse roles of glycine-rich RNA-binding protein 7 in the response of camelina (Camelina sativa) to abiotic stress. Acta Physiol Planta 38:1–11
Wang C, Zhang D-W, Wang Y-C, Zheng L, Yang C-P (2012) A glycine-rich RNA-binding protein can mediate physiological responses in transgenic plants under salt stress. Mol Biol Rep 39:1047–1053
Acknowledgements
We thank Prof. A. Obrępalska-Stęplowska from IPP-INR for her supervision, valuable help and suggestions during the preparation of the manuscript.
Funding
This study was supported by a Grant from the Polish National Science Center (UMO-2014/13/N/NZ9/00703).
Author information
Authors and Affiliations
Contributions
Both authors contributed to the study's conception and design. Material preparation, data collection, and analysis were performed by both authors. The first draft of the manuscript was written by AP and both authors commented on previous versions of the manuscript. Both authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Przybylska, A., Spychalski, M. Changes in the expression level of genes encoding transcription factors and cell wall-related proteins during Meloidogyne arenaria infection of maize (Zea mays). Mol Biol Rep 48, 6779–6786 (2021). https://doi.org/10.1007/s11033-021-06677-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06677-3