Abstract
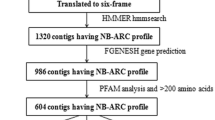
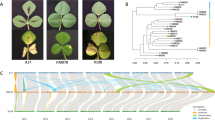
Coriander (Coriandrum sativum L.) is well known vegetable and spice crop grown globally for its leaves and seeds. Stem gall (Protomyces macrosporus L.) is a fungal disease affecting its quality and yield. However, no information is available on SSR markers linked to disease resistance in coriander. Hence, development of co-dominant genetic markers is prerequisite for disease investigations in coriander. In-house stem gall resistance and susceptible cultivars transcriptome data were utilized. Totally, 59,933 and 56,861 transcripts were examined, 9141 and 8346 Simple Sequence Repeats (SSR) were identified and the most abundant type was the tri, followed by di, tetra, penta and hexa nucleotide repeats. A total of ten selected SSR-Functional Domain Markers (FDM) were developed based on functional annotation terms associated with pathogen response and validated among ten coriander cultivars and their transferability was examined in five fennel (Foeniculum vulgare L.) cultivars. Nine primer pairs resulted from amplified bands. Marker ACorSGD-1 shown monomorphic bands among coriander genotypes except Acr-1 showed heterologouse and multiple bands in fennel cultivars. Markers ACorSGD-4, 5, 7 and 9 shown presence in resistant cultivars and absence of bands among susceptible cultivars of coriander and thus, considered to be the candidate markers for disease screening. Marker ACorSGD-6 shown monomorphic bands among coriander. Markers ACorSGD-1, 2, 3, and 5 shown transferability among fennel cultivars. A total of 136 alleles in coriander and fennel were produced. Using UPGMA clustering method a dendrogram was generated and cultivars were grouped into two separate clusters with coriander and fennel. Identified and developed SSR-FDM markers are useful for linkage mapping for disease resistant in coriander.



Similar content being viewed by others
Abbreviations
- SSR:
-
Simple sequence repeats
- FDM:
-
Functional domain marker
- ACorSGD:
-
Ajmer coriander stem gall disease
References
Sharma MM, Sharma RK (2004) Coriander. In: Peter KV (ed) Hand book of herbs and spices, vol 2. Woodhead Publishing Limited, Cambridge
Aruna G, Baskaran V (2010) Comparative study on the levels of carotenoids lutein, zeaxanthin and b-carotene in Indian spices of nutritional and medicinal importance. Food Chem 123:404–409
Choudhary S, Pereira A, Basu S (2014) Effect of cryogenic and conventional grinding on the anti-oxidative potential of coriander and turmeric. Int J Seed Spices 2:85–90
Choudhary S, Pereira A, Basu S, Verma AK (2017) Differential antioxidant composition and potential of some commonly used Indian spices. J Agrisearch 4:160–166
Choudhary S, Jethra G, Sharma R, Verma AK (2017) Microsatellite in coriander: a cross species amplification within Apiaceae family. Int J Curr Microbiol Appl Sci 5:2714–2721
Al-Kordy MA, Abou El-Nasr THS, Sherin AM (2013) Assessing phenotypic and molecular variability in coriander (Coriandrum sativum L.) cultivars. J Appl Sci Res 9:3880–3889
Singh RK, Verma SS, Meena RS, Kumar R (2013) Characterization of coriander (Coriandrum sativum L.) varieties using SDS-PAGE and RAPD markers. Afr J Biotechnol 12(11):1189–1195
Choudhary S, Sharma R, Jethra G, Vishal MK, Tripathi A (2019) Molecular diversity in coriander (Coriandrum sativum) using RAPD and ISSR markers. Indian J Agric Sci 89:193–198
Song X, Wang J, Li N, Ju J, Meng F, Wei C, Liu C, Chen W, Nie F, Zhang Z, Ke G, Li X, Jingjing Hu, Yang Q, Li Y, Li C, Feng S, He G, Yuan J, Pei Q, Tong Yu, Xi K, Zhao W, Lei T, Sun P, Li W, Ge W, Di G, Duan X, Shen S, Cui C, Ying Yu, Xie Y, Zhang J, Hou Y, Wang J, Wang J, Li X-Q, Paterson AH, Wang X (2020) Deciphering the high-quality genome sequence of coriander that causes controversial feelings. Plant Biotechnol J 18:1444–2145
Song X, Nie F, Ma X, Gong K, Yang Q, Wang J, Li N, Sun P, Pei Q, Yu T, Hu J, Li X, Wu T, Feng S, Li X, Wang X (2020) Coriander Genomics Database: a genomic, transcriptomic, and metabolic database for coriander. Hortic Res 7:55
Choudhary S, Naika MBN, Sharma R, Meena RD, Singh R, Lal G (2019) Transcriptome profiling of coriander: a dual purpose crop unravels stem gall resistance genes. J Genet 98:19
Tulsani NJ, Hamid R, Jacob F, Umretiya NG, Nandha AK, Tomar RS, Golakiya BA (2019) Transcriptome landscaping for gene mining and SSR marker development in coriander (Coriandrum sativum L.). Genomics 112(2):1545–1553
Choudhary S, Naika MBN, Meena RD (2020) Identification and expression analysis for candidate genes associated with stem gall disease in Coriander (Coriandrum sativum L.) cultivars. Mol Biol Rep 47:5403–5409
Kalia RK, Rai MK, Kalia S, Singh R, Dhawan AK (2011) Microsatellite markers: an overview of the recent progress in plants. Euphytica 177:309–334
Saha MC, Mian MA, Eujayl I, Zwonitzer JC, Wang L et al (2004) Tall fescue EST-SSR markers with transferability across several grass species. Theor Appl Genet 109(4):783–791
Yu JK, Paik H, Choi JP, Han JH, Choe JK, Hur CG (2010) Functional domain marker (FDM): an in silico demonstration in Solanaceae using simple sequence repeats (SSRs). Plant Mol Biol Rep 28:352–356
Martins WS, Lucas DC, Neves KF, Bertioli DJ (2009) WebSat—a web software for microsatellite marker development. Bioinformation 3(6):282–283
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Rohlf FJ (1997) NTSYS-pc Version. 2.02i numerical taxonomy and multivariate analysis system. Applied Biostatistics Inc., Exeter Software. Setauket, New York
Khare MN, Tiwari SP, Sharma YK (2017) Disease problems in the cultivation of coriander (Coriandrum sativum L.) and their management leading to production of high quality pathogen free seed. Int J Seed Spices 7:1–7
Jain S (2018) In vitro studies on dual culture of Protomyces macrosporus on Coriandrum sativum. Int Res J Nat Appl Sci 5(4):118–132
López PA, Widrlechner MP, Simon PW, Rai S, Boylston TD, Isbell TA, Bailey TB, Gardner CA, Wilson LA (2008) Assessing phenotypic, biochemical, and molecular diversity in coriander (Coriandrum sativum L.) germplasm. Genet Resour Crop Evol 55:247–275
Pareek N, Jakhar M, Malik C (2017) Analysis of genetic diversity in coriander (Coriandrum sativum L.) varieties using random amplified polymorphic DNA (RAPD) markers. J Microbiol Biotechnol Res 1:206–215
Choudhary S, Jethra G, Sharma R, Verma AK (2017) Microsatellite in coriander: a cross species amplification within Apiaceae family. Int J Curr Microbiol Appl Sci 6:2714–2721
Gupta S, Shukla R, Roy S, Sen N, Sharma A (2010) In silico SSR and FDM analysis through EST sequences in Ocimum basilicum. POJ 3(4):121–128
Sahu J, Sarmah R, Dehury B, Sarma K, Sahoo S, Sahu M, Barooah M, Modi MK, Sen P (2012) Mining for SSRs and FDMs from expressed sequence tags of Camellia sinensis. Bioinformation 8(6):260–266
Sorkheh K, Prudencio AS, Ghebinejad A et al (2016) In silico search, characterization and validation of new EST-SSR markers in the genus Prunus. BMC Res Notes 9:336
Ray M, Pradhan SP, Sahoo S (2019) Computational in sight into identification and analysis of SSR-FDM in Citrus limon. Int J Plant Biol Res 7(1):1111
Singh HB, Singh A, Tripathi A, Rai SK, Katiyar RS, Johri JK, Singh SP (2003) Evaluation of coriander germplasms for stem gall disease. Genet Resour Crop Evol 50(4):339–343
Malhotra SK, Kakani RK, Sharma YK, Singh DK (2016) Ajmer coriander-1 (NRCSS, ACR-1), resistant to stem gall disease—an innovative farming technology. Indian J Arecanut Spices Med Plants 18(3):3–7
Wei K, Chen J, Wang Y, Chen Y, Chen S, Lin Y, Pan S, Zhong X, Xie D (2012) Genome-wide analysis of bZIP-encoding genes in maize. DNA Res 19:463–476
Häffner E, Konietzki S, Diederichsen E (2015) Keeping control: the role of senescence and development in plant pathogenesis and defense. Plants 4:449–488
Jethra G, Choudhary S, Sharma V (2018) Development and characterization of novel set of polymorphic SSR markers for Dill (Anethum graveolens L.). Ann Plant Sci 7(6):2347–2351
Jethra G, Choudhary S, Sharma V (2020) Identification and characterization of microsatellite markers in fenugreek: an inter-family amplification. Legume Res 43:611–616
Acknowledgements
We thank Director, NRCSS for timely support and encouragement during the research work. The research is supported by the ICAR-NRC on Seed Spices, Ajmer, Rajasthan, India. Authors also acknowledge the support received by the UHS, Bagalkot, Karnataka for infrastructure facilities.
Author information
Authors and Affiliations
Contributions
Conceptualization and methodology developed: SC, MBNN, Formal analysis and investigation: SC, MBNN and RDM Writing—original draft preparation: MBNN, SC and RDM.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Our research does not include Human Participants and/or Animals.
Consent to participate
Our research does not include Human and/or Animals participants.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Choudhary, S., Naika, M.B.N. & Meena, R.D. Development and characterization of genic SSR-FDM for stem gall disease resistance in coriander (Coriandrum sativum L.) and its cross species transferability. Mol Biol Rep 48, 3963–3970 (2021). https://doi.org/10.1007/s11033-021-06396-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06396-9