Abstract
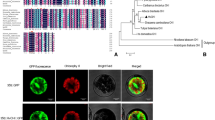
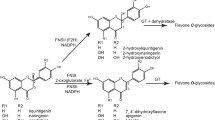
Camellia nitidissima Chi. is an ornamental plant of the genus Camellia L. Its flowers contain a lot of flavonoids and polyphenols. Flavonoid 3′-hydroxylase (F3′H) plays an important role in the synthesis of flavonoids, polyphenols and anthocyanins. We used PCR amplification, quantitative PCR, High-performance liquid chromatography, subcellular localization, and agrobacterium-mediated leaf disk method to study the the function of CnF3′H. The full length of CnF3′H was 1859 bp (GenBank code: HQ290518.1), with an open reading frame of 1577 bp, and encoded 518 amino acid. A phylogenetic tree analysis showed that CnF3′H was closely related to Camellia sinensis L. and C. sinensis cultivar Zhonghuang. CnF3′H was expressed in flowers, leaves, fruits, sepals, petals and stamens of C. nitidissima, and during the flowering process the expression level in flower decreased initially and then increased. CnF3′H expression was significantly positive correlated with polyphenol contents in C. nitidissima. A CnF3′H-EGFP expression vector was constructed to do the subcellular localization, we found that CnF3′H was obviously localized in the nuclear envelope and cytomembrane. In transgenic tobacco flowers, the total polyphenol content and various polyphenol constituents were significantly increased with high CnF3′H expression level, while total flavonoid contents and some flavonol constituents were increased slightly. These findings suggest that CnF3′H promotes the synthesis of polyphenols better than flavonoids.





Similar content being viewed by others
Data Availability
All data are presented in the manuscript. Camellia nitidissima Chi. tissues were collected from the National Camellia Germplasm Resource Bank (Guangxi, China).
References
Zhang H, Ren S (1998) Flora of China (Theaceae). In: Zhang H (ed) Flora Reipublicae popularis Sinicae, vol 49. Science Press, Beijing, pp 1–194
Jiang L, Li J, Tong R, He L, Zhang L, Li Z, Huang X (2019) Relationship between flower color and important cellular environment elemental factors in yellow Camellia. Guihaia 39(12):1605–1612. https://doi.org/10.11931/guihaia.gxzw201809019
Li X, Wang J, Sun Z, Wang J, Yin H, Fan Z, Li J (2019) Flavonoid components in flowers from three species of section Chrysantha Chang in Camellia. Guihaia 39(7):917–924. https://doi.org/10.11931/guihaia.gxzw201809009
Mouradov A, Spangenberg G (2014) Flavonoids: a metabolic network mediating plants adaptation to their real estate. Front Plant Sci 5:620. https://doi.org/10.3389/fpls.2014.00620
Passat DN (2012) Interactions of black and green tea water extracts with antibiotics activity in local urinary isolated Escherichia coli. J Al-Nahrain Univ 15(3):134–142. https://doi.org/10.22401/JNUS.15.3.19
Tanikawa N, Kashiwabara T, Hokura A, Abe T, Shibata M, Nakayama M (2008) A peculiar yellow flower coloration of Camellia using aluminum-flavonoid interaction. J Jpn Soc Hortic Sci 77(4):402–407. https://doi.org/10.2503/jjshs1.77.402
Chapple C (1998) Molecular-genetic analysis of plant cytochrome P450-dependent monooxygenases. Annu Rev Plant Biol 49(1):311–343. https://doi.org/10.1146/annurev.arplant.49.1.311
Hou J, Tong L, Cui G, Xu Z, Li Y (2011) Research advances of plant flavonoid 3′-hydroxylase (F3′H) gene. Plant Physiol J 47:641–647
Nakatsuka T, Abe Y, Kakizaki Y, Yamamura S, Nishihara M (2007) Production of red-flowered plants by genetic engineering of multiple flavonoid biosynthetic genes. Plant Cell Rep 26(11):1951–1959. https://doi.org/10.1007/s00299-007-0401-0
Nakatsuka T, Sasaki N, Nishihara M (2014) Transcriptional regulators of flavonoid biosynthesis and their application to flower color modification in Japanese gentians. Plant Biotechnol 31:389–399. https://doi.org/10.5511/plantbiotechnology.14.0731a
Grotewold E (2006) The genetics and biochemistry of floral pigments. Annu Rev Plant Biol 57:761–780. https://doi.org/10.1146/annurev.arplant.57.032905.105248
Jia Z (2017) Studies on roles of flavonoid quercetin and transcription factor ATMYB44 in induction and regulation of defense responses in Arabidopsis thaliana. Nanjing Agricultural University, Nanjing. https://doi.org/10.7666/d.Y1986714
Brugliera F, Barri-Rewell G, Holton TA, Mason JG (1999) Isolation and characterization of a flavonoid 3′-hydroxylase cDNA clone corresponding to the Ht1 locus of Petunia hybrida. Plant J 19(4):441–451. https://doi.org/10.1046/j.1365-313X.1999.00539.x
Schoenbohm C, Martens S, Eder C, Forkmann G, Weisshaar B (2000) Identification of the Arabidopsis thaliana flavonoid 3′-hydroxylase gene and functional expression of the encoded P450 enzyme. Biol Chem 381(8):749–753. https://doi.org/10.1515/BC.2000.095
Sirim D, Wagner F, Lisitsa A, Pleiss J (2009) The cytochrome P450 engineering database: integration of biochemical properties. BMC Biochem. https://doi.org/10.1186/1471-2091-10-27
Han Y, Vimolmangkang S, Soria-Guerra RE, Rosales-Mendoza S, Zheng D, Lygin AV, Korban SS (2010) Ectopic expression of apple F3′H genes contributes to anthocyanin accumulation in the Arabidopsis tt7 mutant grown under nitrogen stress. Plant Physiol 153(2):806–820. https://doi.org/10.1104/pp.109.152801
Li L, Cheng H, Chen X, Cheng S (2015) Molecular cloning, characterization and expression of flavonoid 3′hydroxylase-like protein gene from Ginkgo biloba. Acta Hortic Sin 42:643–654
Zhou TS, Zhou R, Yu YB, Xiao Y, Li DH, Xiao B, Yu O, Yang YJ (2016) Cloning and characterization of a flavonoid 3′-hydroxylase gene from tea plant (Camellia sinensis). Int J Mol Sci 17(2):261. https://doi.org/10.3390/ijms17020261
Huang M, Wen Z, Chi Y, Peng Z, Chen Q (2017) Molecular cloning and expression analysis of flavonoids 3′-hydroxylase (CaF3′H) in Canarium album. Mol Plant Breed 15(3):839–847
Lukačin R, Urbanke C, Gröning I, Matern U (2000) The monomeric polypeptide comprises the functional flavanone 3β-hydroxylase from Petunia hybrida. FEBS Lett 467(2–3):353–358. https://doi.org/10.1016/s0014-5793(00)01116-9
Masukawa T, Cheon K-S, Mizuta D, Kadowaki M, Nakatsuka A, Kobayashi N (2019) Development of mutant RsF3′H allele-based marker for selection of purple and red root in radish (Raphanus sativus L. var. longipinnatus LH Bailey). Euphytica 215(7):119. https://doi.org/10.1007/s10681-019-2442-1
Masukawa T, Cheon K-S, Mizuta D, Nakatsuka A, Kobayashi N (2018) Insertion of a retrotransposon into a flavonoid 3′-hydroxylase homolog confers the red root character in the radish (Raphanus sativus L. var. longipinnatus LH Bailey). Hortic J 87(1):89–96. https://doi.org/10.2503/hortj.OKD-075
Nakatsuka T, Nishihara M, Mishiba K, Yamamura S (2006) Heterologous expression of two gentian cytochrome P450 genes can modulate the intensity of flower pigmentation in transgenic tobacco plants. Mol Breed 17(2):91–99. https://doi.org/10.1007/s11032-005-2520-z
Tanaka M, Sakai T, Takahata Y (2019) Allele dosage-dependent selection of recessive F3′H allele homozygote altered anthocyanin composition in sweetpotato. Mol Breed 39(11):1–12. https://doi.org/10.1007/s11032-019-1062-8
Zhou X (2012) Clonging and function reasearch of pigment genes from Camallia nitidissma. Chinese Academy of Forestry Sciences, Beijing. https://doi.org/10.7666/d.D603419
Olsen KM, Hehn A, Jugdé H, Slimestad R, Larbat R, Bourgaud F, Lillo C (2010) Identification and characterisation of CYP75A31, a new flavonoid 3′5′-hydroxylase, isolated from Solanum lycopersicum. BMC Plant Biol 10(1):21. https://doi.org/10.1186/1471-2229-10-21
McGinnis S, Madden TL (2004) BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. https://doi.org/10.1093/nar/gkh435
Wilkins MR, Gasteiger E, Gooley AA, Herbert BR, Molloy MP, Binz P-A, Ou K, Sanchez J-C, Bairoch A, Williams KL (1999) High-throughput mass spectrometric discovery of protein post-translational modifications. J Mol Biol 289(3):645–657. https://doi.org/10.1006/jmbi.1999.2794
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680. https://doi.org/10.1093/nar/22.22.4673
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Jiang L, Fan Z, Tong R, Zhou X, Li J, Yin H (2020) Functional diversification of the dihydroflavonol 4-reductase from Camellia nitidissima Chi. in the control of polyphenol biosynthesis. Genes 11(11):1341. https://doi.org/10.3390/genes11111341
Gelvin SB (2003) Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol Mol Biol Rev 67(1):16–37. https://doi.org/10.1128/MMBR.67.1.16-37.2003
Ma L, Lukasik E, Gawehns F, Takken FL (2012) The use of agroinfiltration for transient expression of plant resistance and fungal effector proteins in Nicotiana benthamiana leaves. Methods Mol Biol 835:61–74. https://doi.org/10.1007/978-1-61779-501-5_4
Yamaguchi M, Sasaki T, Sivaguru M, Yamamoto Y, Osawa H, Ahn SJ, Matsumoto H (2005) Evidence for the plasma membrane localization of Al-activated malate transporter (ALMT1). Plant Cell Physiol 46(5):812–816. https://doi.org/10.1093/pcp/pci083
He L, Zhao S, Hu Z (2008) Gene and function research progress of plant cytochrome P450s. Pharm Biotechnol 15(2):142. https://doi.org/10.3724/SP.J.1005.2008.01026
Seitz C, Ameres S, Forkmann G (2007) Identification of the molecular basis for the functional difference between flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase. FEBS Lett 581(18):3429–3434. https://doi.org/10.1016/j.febslet.2007.06.045
Murakami K, Mihara K, Omura T (1994) The transmembrane region of microsomal cytochrome P450 identified as the endoplasmic reticulum retention signal. J Biochem 116(1):164–175. https://doi.org/10.1016/0165-022X(94)90059-0
Yamazaki S, Sato K, Suhara K, Sakaguchi M, Mihara K, Omura T (1993) Importance of the proline-rich region following signal-anchor sequence in the formation of correct conformation of microsomal cytochrome P-450s. J Biochem 114(5):652–657. https://doi.org/10.1007/s00601-013-0691-4
He F, Chen WK, Yu KJ, Ji XN, Duan CQ, Reeves MJ, Wang J (2015) Molecular and biochemical characterization of the UDP-glucose: anthocyanin 5-O-glucosyltransferase from Vitis amurensis. Phytochemistry 117:363–372. https://doi.org/10.1016/j.phytochem.2015.06.023
Wang H, Wang W, Zhang P, Pan Q, Zhan J, Huang W (2010) Gene transcript accumulation, tissue and subcellular localization of anthocyanidin synthase (ANS) in developing grape berries. Plant Sci 179(1–2):103–113. https://doi.org/10.1016/j.plantsci.2010.04.002
Li H, Liu J, Jin W, Liang Z (2019) Cloning, subcellular localization and spatio-temporal expression analysis of a flavonoid 3-O-glucosyltransferase gene (SmUF3GT) in Salvia miltiorrhiza. China J Chin Mater Med 44(10):2038–2045. https://doi.org/10.19540/j.cnki.cjcmm.20190301.009
Toda K, Yang D, Yamanaka N, Watanabe S, Harada K, Takahashi R (2002) A single-base deletion in soybean flavonoid 3′-hydroxylase gene is associated with gray pubescence color. Plant Mol Biol 50(2):187–196. https://doi.org/10.1023/A:1016087221334
Stafford H (1974) Possible multienzyme complexes regulating the formation of C6–C3 phenolic compounds and lignins in higher plants. Recent Adv Phytochem 8:53–79. https://doi.org/10.1016/B978-0-12-612408-8.50009-3
Jiang X, Liu Y, Li W, Zhao L, Meng F, Wang Y, Tan H, Yang H, Wei C, Wan X (2013) Tissue-specific, development-dependent phenolic compounds accumulation profile and gene expression pattern in tea plant [Camellia sinensis]. PLoS ONE. https://doi.org/10.1371/journal.pone.0062315
He H, Pan H, Zhang N, He L (2015) Chromolaena odorata Flavonoid 3′-hydroxylase gene cloning and its expression in tobacco. Acta Agron Sin 41(3):479–486. https://doi.org/10.3724/SP.J.1006.2015.00479
Zhang S, Guo H, Pei X, Li C, Cheng H (2009) Expression of flavonoid 3′-hydrolyase from Eupatorium adenophorum in tobacco. Sci Agric Sin 42(12):4182–4186. https://doi.org/10.3864/j.issn.0578-1752.2009.12.008
Acknowledgements
The authors thank the members of the National Forestry and Grassland Administration Economic Forest Product Quality Supervision, Inspection and Testing Center (Hangzhou) for their assistance in condition establishment and detection of high-performance liquid phase.
Funding
This work was supported by the National Key R&D Program of China (2019YFD1001005), the special funds for basic scientific research expenses of public welfare research institutes of the Chinese academy of forestry (CAFYBB2017ZF001), The National key projects for international scientific and technological innovation cooperation among governments (2016YFE0126100), The National Natural Science Foundation of China (No. 31860228), the Key projects of Natural Science Foundation of the Guangxi Zhuang Autonomous Region (2018GXNSFDA281007), and Scientific Research Foundation of Yulin Normal University for high-level talents (G2018025).
Author information
Authors and Affiliations
Contributions
JL finished the main experimental contentc, wrote and modified the paper. FZ and TR participated in the experiment and data collection together. ZX provided the foundation and thought for the preliminary study. LJ was the architect and director of the project. YH was responsible for the experimental design, and guided the writing and modification of the paper. All authors read and agree to the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Jiang, L., Fan, Z., Tong, R. et al. Flavonoid 3′-hydroxylase of Camellia nitidissima Chi. promotes the synthesis of polyphenols better than flavonoids. Mol Biol Rep 48, 3903–3912 (2021). https://doi.org/10.1007/s11033-021-06345-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06345-6