Abstract
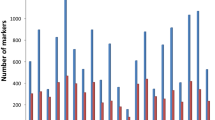
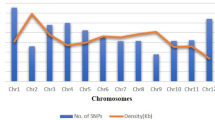
Safflower (Carthamus tinctorius L.) is one of the most important oilseed crops for its seed oil rich in unsaturated fatty acids. Precise utilization of diverse genetic resources is fundamental in breeding programs to improve high yield genotypes with desirable traits. In this study, for the first time we report successful application of DArTseq technology; an efficient genotyping-by-sequencing (NGS); to analysis genetic diversity and population structure of 89 safflower accessions from worldwide origins. Totally, 19,639 DArTseq markers (10,130 SilicoDArTs and 9509 SNPs) generated through DArTseq genotyping. After filtering the data, 3431 polymorphic DArTseq markers (1136 SilicoDArTs and 2295 SNPs) used for genetic diversity, population structure and linkage disequilibrium analysis in safflower genotypes. All the SilicoDArT and SNP markers showed high reproducibility and call rate. Polymorphism information content (PIC) values ranged from 0.1 to 0.5, while ≥ 0.50% of SilicoDArTs and ≥ 0.64% SNPs showed PIC values more than median. Genotypes grouping using DArTseq markers resulted in three distinct clusters. Results showed weak correlation between safflower diversity pattern and origins. Analysis of molecular variance revealed that the majority of genetic variation was attributed to the differences among varieties within cluster populations and there was no significant molecular variance between origins. However, safflower of accessions belonged to Iran, Turkey, Pakistan and India indeed appear to be genetically similar and grouped close in referred cluster, while the accessions from Near East (Afghanistan, China) being distinct. Our results were in agreement with hypothesis that safflower domesticated in somewhere west of Fertile Crescent and then expanded through Africa and Europe. Present study using a panel of globally diverse safflower accessions and large number of DArTseq markers set the stage for future analysis of safflower domestication using large germplasm from proposed domestication centers. Also, studied germplasm in this study can be used as a valuable source for future genomic studies in safflower for mapping desirable traits through genome-wide association mapping studies.











Similar content being viewed by others
Abbreviations
- NGS:
-
Next generation sequencing
- DArT:
-
Diversity array technology
- SNP:
-
Single nucleotide polymorphism
- DAPC:
-
Discriminant analysis of principal component
- LD:
-
Linkage disequilibrium
References
Asp ML, Collene AL, Norris LE, Cole RM, Stout MB, Tang SY, Hsu JC, Belury MA (2011) Time-dependent effects of safflower oil to improve glycemia, inflammation and blood lipids in obese, post-menopausal women with type 2 diabetes: a randomized, double-masked, crossover study. Clin Nutr 30:443–449
Kumar S, Ambreen H, Murali TV, Bali S, Agarwal M, Kumar A, Goel S, Jagannath A (2015) Assessment of genetic diversity and population structure in a global reference collection of 531 accessions of Carthamus tinctorius L. (Safflower) using AFLP markers. Plant Mol Biol Rep 33:1299–1313
Burke JM, Tang S, Knapp SJ, Rieseberg LH (2002) Genetic analysis of sunflower domestication. Genetics 161:1257–1267
Kumar S, Ambreen H, Variath MT, Rao AR, Agarwal M, Kumar A, Goel S, Jagannath A (2016) Utilization of molecular, phenotypic and geographical diversity to develop compact composite core collection in the oilseed crop, Safflower (Carthamus tinctorius L.) through maximization strategy. Front Plant Sci 7:1554
Norris LE, Collene AL, Asp ML, Hsu JC, Liu LF, Richardson JR, Li D, Bell D, Osei K, Jackson RD, Belury MA (2009) Comparison of dietary conjugated linoleic acid with safflower oil on body composition in obese postmenopausal women with type 2 diabetes mellitus. Am J Clin Nutr 90:468–476
Knowles P, Ashri A (1995) Safflower—Carthamus Tinctorius (Compositae): In evolution of crop plants. Ed by Smartt J, Simmonds NW. Harlow, UK: Longman; p 47–50.
Knowles PF (1969) Centers of plant diversity and conservation of crop germplasm- Safflower. Econ Bot 23(4):324–329
Amini F, Saeidi G, Arzani A (2008) Study of genetic diversity in safflower genotypes using agro-morphological traits and RAPD markers. Euphytica 163:21–30
Khan MA, von Witzke-Ehbrecht S, Maass BL, Becker HC (2009) Relationships among different geographical groups, agro-morphology, fatty acid composition and RAPD marker diversity in safflower (Carthamus tinctorius). Genet Resour Crop Evol 56:19–30
Talebi R, Nosrati S, Etminan A, Naji AM (2018) Genetic diversity and population structure analysis of landrace and improved safflower (Cartamus tinctorious L.) germplasm using arbitrary functional gene-based molecular markers. Biotech Biotech Equip 32:1183–1194
Golkar P, Mokhtari N (2018) Molecular diversity assessment of a world collection of safflower genotypes by SRAP and SCoT molecular markers. Physiol Mol Biol Plants 24(6):1261–1271
Brown AHD (1989) Core collections—a practical approach to genetic-resources management. Genome 31(2):818–824
Johnson RC, Stout DM, Bradley VL (1993) "The US collection: a rich source of safflower germplasm", In: Dajue L, Yuanzhou H (eds) Proceedings of the third international safflower conference, Beijing: Beijing Botanical Garden; Institute of Botany, Chinese Academy of Sciences, pp 202–208.
Dwivedi SL, Upadhyaya HD, Hegde DM (2005) Development of core collection using geographic information and morphological descriptors in safflower (Carthamus tinctorius L.) germplasm. Genet Resour Crop Evol 52:821–830
Pearl SA, Burke JM (2014) Genetic diversity in Carthamus tinctorius (Asteraceae; safflower), an underutilized oilseed crop. Am J Bot 101:1640–1650
Golkar P, Arzani A, Rezaei AM (2011) Genetic variation in safflower (Carthamus tinctorious L.) for seed quality-related traits and inter-simple sequence repeat (ISSR) markers. Int J Mol Sci 12:2664–2677
Kiran BU, Mukta N, Kadirvel P, Alivelu K, Senthilvel S, Kishore P, Varaprasad KS (2017) Genetic diversity of safflower (Carthamus tinctorius L.) germplasm as revealed by SSR markers. Plant Genet Resour 15(1):1–11
Ambreen H, Kumar S, Kumar A, Agarwal M, Jagannath A, Goel S (2018) Association mapping for important agronomic traits in safflower (Carthamus tinctorius L.) core collection using microsatellite markers. Front Plant Sci 9:402
Alam M, Neal J, O’Connor K, Kilian A, Topp B (2018) Ultra-high-throughput DArTseq-based silicoDArT and SNP markers for genomic studies in macadamia. PLoS ONE 13(8):e0203465
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6:e19379
Beattie WG, Meng L, Turner SL, Varma RS, Dao DD, Beattie KL (1995) Hybridization of DNA targets to glass-tethered oligonucleotide probes. Mol Biotechnol 4(3):213–225
Bowers JE, Bachlava E, Brunick RL, Rieseberg LH, Knapp SJ, Burke JM (2012) Development of a 10,000 locus genetic map of the sunflower genome based on multiple crosses. G3 (Bethesda) 2(7):721–729
Bowers JE, Pearl SA, Burke JM (2016) Genetic mapping of millions of SNPs in safflower (Carthamus tinctorius L) via Whole-Genome Resequencing. G3 (Bethesda) 6(7):2203–2211
Truco MJ, Ashrafi H, Kozik A, van Leeuwen H, Bowers J, Wo SR, Stoffel K, Xu H, Hill T, Van Deynze A, Michelmore RW (2013) An ultra-high-density, transcript-based, genetic map of Lettuce. G3 (Bethesda) 3(4):617–631
Chapman MA, Burke JM (2007) DNA sequence diversity and the origin of cultivated safflower (Carthamus tinctorius L.; Asteraceae). BMC Plant Biol 7:60
Akbari M, Wenzl P, Caig V, Carling J, Xia L, Yang S, Uszynski G, Mohler V, Lehmensiek A, Kuchel H, Hayden MJ, Howes N, Sharp P, Vaughan P, Rathmell B, Huttner E, Kilian A (2006) Diversity arrays technology (DArT) for high-throughput profiling of the hexaploid wheat genome. Theor Appl Genet 113:1409–1420
Nadeem MA, Habyarimana E, Ciftci V, Nawaz MA, Karakoy T, Comertpay G, Shahid MQ, Hatipoğlu R, Yeken MZ, Ali F, Ercişli S, Gyuhwa Chung G, Baloch FS (2018) Characterization of genetic diversity in Turkish common bean gene pool using phenotypic and whole-genome DArTseq-generated silicoDArT marker information. PLoS ONE 13(10):e0205363
Ates D, Asciogul TK, Nemli S, Erdogmus S, Esiyok D, Tanyolac MB (2018) Association mapping of days to flowering in common bean (Phaseolus vulgaris L.) revealed by DArT markers. Mol Breed 38:113
Kilian A, Wenzl P, Huttner E, Carling J, Xia L, Blois H, Caig V, Heller-Uszynsk K, Jaccoud D, Hopper C, Aschenbrenner-Kilian M, Evers M, Peng K, Cayla C, Hok P, Uszynski G (2012) Diversity arrays technology: a generic genome profiling technology on open platforms. Meth Mol Biol 888:67–89
Edet OU, Gorafi YSA, Cho SW, Kishii M, Tsujimoto H (2018) Novel molecular marker-assisted strategy for production of wheat–Leymus mollis chromosome addition lines. Sci Rep 8:16117
Ndjiondjop MN, Semagn K, Gouda AC, Kpeki SB, Dro Tia D, Sow M, Goungoulou A, Sie M, Perrier X, Ghesquiere A, Warburton ML (2017) Genetic variation and population structure of Oryza glaberrima and development of a mini-core collection using DArTseq. Front Plant Sci 8:1748
Pailles Y, Ho S, Pires IS, Tester M, Negrão S, Schmöckel SM (2017) Genetic diversity and population structure of two tomato species from the Galapagos Islands. Front Plant Sci 8:138
Zaitoun SYA, Jamous RM, Shtaya MJ, Mallah OB, Eid IS, Ali-Shtayeh MS (2018) Characterizing Palestinian snake melon (Cucumis melo var. flexuosus) germplasm diversity and structure using SNP and DArTseq markers. BMC Plant Biol 18:246
Lassner MW, Peterson P, Yoder JI (1989) Simultaneous amplification of multiple DNA fragments by polymerase chain reaction in the analysis of transgenic plants and their progeny. Plant Mol Biol Rep 7:116–128
Talebi R (2008) An alternative strategy in rapid DNA extraction protocol for high throughput RAPD analysis in chickpea and its wild related species. J Appl Biol Sci 2(3):121–124
Monostori I, Szira F, Tondelli A, Arendas T, Gierczik K, Cattivelli L, Galiba G, Vagujfalvi A (2017) Genome-wide association study and genetic diversity analysis on nitrogen use efficiency in a Central European winter wheat (Triticum aestivum L.) collection. PLoS ONE 12(12):e0189265
Baloch FS, Alsaleh A, Shahid MQ, Çiftçi V, Sáenz de Miera L, Aasim M, Nadeem MA, Aktas S, Ozkan H, Hatipoglu RA (2017) whole genome DArTseq and SNP analysis for genetic diversity assessment in durum wheat from central fertile crescent. PLoS ONE 12(1):e0167821
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Cruz VMV, Kilian A, Dierig DA (2013) Development of DArT marker platforms and genetic diversity assessment of the US collection of the new oilseed crop lesquerella and related species. PLoS ONE 8:e64062
Perrier X, Flori A, Bonnot F (2003) Data analysis methods: In: Hamon, P., Seguin, M., Perrier, X., Glaszmann, J.C. Genetic diversity of cultivated tropical plants. Science Publishers, Enfield, 43–76.
Perrier X, Jacquemoud-Collet JP (2006) DARwin software, https://darwin.cirad.fr/darwin.
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTU RE: a simulation study. Mol Ecol 14:2611–2620
Earl DA, vonholdt BM, (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTU RE output and implementing the Evanno method. Conserv Genet Resour 4(2):359–361
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11(1):94
Core Team R (2014) R: a language and environment for statistical computing. R Core Team, Vienna
Nielsen NH, Backes G, Stougaard J, Andersen SU, Jahoor A (2014) Genetic diversity and population structure analysis of European hexaploid bread wheat (Triticum aestivum L.) varieties. PLoS ONE 9:e94000
Chapman MA, Hvala J, Strever J, Burke JM (2010) Population genetic analysis of safflower (Carthamus tinctorius; Asteraceae) reveals a Near Eastern origin and five centers of diversity. Am J Bot 97:831–840
Talebi R, Abhari SA (2016) Evaluation of genetic diversity in safflower (Carthamus tinctorius L.) using agro-morphological, fatty acid composition and ISSR molecular markers. Res J Biotech 11(7):19–27
Lee GA, Sung JS, Lee SY, Chung JW, Yi JY, Kim YY, Lee MC (2014) Genetic assessment of safflower (Carthamus tinctorius L.) collection with microsatellite markers acquired via pyrosequencing method. Mol Ecol Resour 14:69–78
Fayaz F, Aghaee M, Talebi R, Azadi A (2019) Genetic diversity and molecular characterization of Iranian durum wheat landraces (Triticum turgidum durum (Desf) Husn) using DArT markers. Biochem Genet 57:98–116
Wambugu PW, Ndjiondjop MN, Henry RJ (2018) Role of genomics in promoting the utilization of plant genetic resources in genebanks. Brief Funct Genom 17:198–206
Onda Y, Mochida K (2016) Exploring genetic diversity in plants using high-throughput sequencing techniques. Curr Genom 17:358–367
Ashri A (1975) Evaluation of the germplasm collection of safflower, Carthamus tinctorius L. V. Distribution and regional divergence for morphological characters. Euphytica 24:651–659
Pearl SA, Bowers JE, Reyes-Chin-Wo S, Michelmore RW, Burke JM (2014) Genetic analysis of safflower domestication. BMC Plant Biol 14:43
Acknowledgements
This study was supported by the Islamic Azad University, Sanandaj Branch, Iran and Sararood Dryland Agricultural Research Institute, Kermanshah, Iran.
Funding
The author(s) received no financial support for the research, authorship, and/or publication of this article.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
No ethical issues were promulgated.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11033_2020_5312_MOESM2_ESM.xlsx
Supplementary file2 Kinship matrix for pairs of genotypes based on SilicoDArT and SNP marker calculated in TASSEL (XLSX 233 kb)
Rights and permissions
About this article
Cite this article
Hassani, S.M.R., Talebi, R., Pourdad, S.S. et al. In-depth genome diversity, population structure and linkage disequilibrium analysis of worldwide diverse safflower (Carthamus tinctorius L.) accessions using NGS data generated by DArTseq technology. Mol Biol Rep 47, 2123–2135 (2020). https://doi.org/10.1007/s11033-020-05312-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05312-x