Abstract
A conversion of amyloplasts into chloroplasts in the potato tuber after light exposure is known as tuber greening and is one of the major causes of tuber loss. We report here the first mapping of the factors affecting tuber greening in potato. We used an F1 mapping population of diploid potatoes and DArTseq™ markers to construct a genetic map. The individuals of the mapping population, parents and standards were phenotyped for two tuber greening parameters: external tuber greening and internal greening depth on 0–5 scales in three years 2015, 2016 and 2018. The results were used for the analysis of Quantitative Trait Loci (QTLs) by an interval QTL mapping. Two most important QTLs were covering large regions of chromosomes VII and X and had the strongest effect on both greening parameters in data sets obtained in particular years and in the mean data set. Variance observed in the mean tuber greening could be ascribed in 16.9% to the QTL on chromosome VII and in 23.4% to the QTL on chromosome X. The QTL on chromosome VII explained 13.1%, while the QTL on chromosome X explained up to 17.7% of the variance in the mean tuber greening depth. Additional, minor QTLs were year- and/or trait-specific. The QTLs on chromosomes VII and X determine big parts of the observed tuber greening variation and should be investigated further in order to identify the genes underlying their effects but also should be taken into account when selecting non-greening potato lines in the breeding process.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
According to Food and Agriculture Organization of the United Nations approximately 1/3 (1.3 billion tons) of the globally produced food is lost or wasted each year. Food losses and waste of root and tuber crops, such as potato (Solanum tuberosum L.), exceed that average and are estimated at 40–50% (FAO Key Facts). Potato is the third most important food crop for human consumption and with global annual production of 388 million tons (estimation from 2017, FAO [1]) it feeds over a billion people.
Potato tuber greening is one of the major causes of tuber quality loss and rejection by a consumer and processing industry. Annual losses due to tuber greening have been estimated at 14–17% of crop value in USA [2] and 17% of fresh potato produce in South Africa [3]. Tuber greening is a phenomenon of conversion of amyloplasts into chloroplasts just beneath the tuber periderm resulting from exposure of tubers to light [4]. Light exposure can also lead to toxic glycoalkaloids synthesis and accumulation in the periderm via pathway independent from chlorophylls synthesis [5]. Even though tuber greening and glycoalkaloids content are not necessarily correlated, green tubers are rejected due to reduced visual appeal but also due to the potentially increased glycoalkaloids content resulting from light exposure of which greening is a clear indicator. This is perhaps, an unnecessary precaution as research has shown that regardless of greening level, concentrations of total glycoalkaloids in the flesh samples of four potato cultivars remained within limits presumed safe for human consumption [6]. Greening can occur at any stage of the supply chain from the field to the household storage and it is a challenge for growers, retailers and consumers. To reduce losses caused by potato tuber greening, a number of factors can be managed. Those factors have been listed and divided into pre- and post-harvest ones in a recent review by Tanios et al. [4]. Among the pre-harvest factors affecting tuber greening, there were mentioned: cultivar’s genetic predisposition, planting depth, nitrogen fertilization (increased soil nitrogen rates enhance greening) and tuber maturity. The post-harvest factors included storage conditions and duration, as well as handling of tubers after storage (light conditions, packaging etc.). Coating with oils, waxes and surfactants as well as chemical and irradiation treatments of potato tubers have been shown to have a potential to decrease greening tendency.
Growing cultivars resistant to tuber greening is a simple and cost-efficient approach as it does not require any modifications of the growing, storage and packaging procedures, or additional tuber treatments. The fact that the tendency for tuber greening is genetically determined has been described previously [7,8,9]. By analyses of progenies of crosses between greening and ‘non-greening’ potato cultivars Akeley et al. [7] have shown that tuber greening is a polygenic trait with incomplete dominance. The quantitative character of the trait has been confirmed by Parfitt and Peloquin [9] using diploid potato families. Reeves [10] has measured three components of greening (external colour, internal colour and depth of colour) in potato cultivars with different skins (russet, red or white) and demonstrated that there was variation in tendency for greening in the three groups of cultivars. However, cultivars with russet skin showed smaller depth of greening as skin types confound resistance due to presence of light adsorbing anthocyanin pigments and opaque russeting. The three components of greening appeared to be independent [10]. In contrary, Jakuczun and Zimnoch-Guzowska [11] have found significant and relatively high correlations between external greening and depth of tuber greening when studying 17 unselected diploid potato families, although they have suggested that the two studied greening components might have been inherited partially independently. Valuable source of resistance to tuber greening has been identified in wild species Solanum microdontum using a different approach [12]. The authors scored tuber greening as a qualitative trait (white or green) and found 10% of S. microdontum families were ‘white’ while none of the 185 North American and European potato cultivars was scored in this category. However, within progenies from crosses ‘white’ × ‘white’, the authors noted variation in tendencies for tuber greening and presence of individuals with green, light green, white and ultra-white tubers after four days of exposure to light typical for supermarkets. Only in three out of 27 such families all the individuals were scored as ‘white’ or ‘ultra-white’, while the progenies from ‘green’ × ‘green’ crosses had consistently strong tendencies for tuber greening [12]. This wild species has been successfully exploited in the introgression into cultivated potato genepool for pre-breeding purposes aiming at greening-resistant stocks [13]
As postulated by Tanios et al. [4], “there is strong evidence that breeding for resistance is possible”, and “an improved understanding of the genetic basis of tuber greening will greatly assist in the identification of molecular markers for genes or regulatory elements associated with reduced greening and thus, accelerate the breeding of cultivars with low tendency to accumulate chlorophyll”. The goal of our study was to perform the first mapping of the genetic factors affecting tuber greening in potato genome. We used an F1 mapping population of diploid potatoes resulting from a cross between parents differing in greening tendency and DArTseq™ markers to construct a genetic map. The individuals of the mapping population, parents and standards were phenotyped for two tuber greening parameters: external tuber greening and internal greening depth on 0–5 scales. The results were used for the analysis of Quantitative Trait Loci (QTLs) that improved our understanding of the tuber greening genetics and that can serve for development of genetic markers for assisting the breeding of potato cultivars resistant to tuber greening.
Material and methods
Plant material
Plant material was a diploid F1 mapping population SEN 12-01 (176 individuals). This population was previously used for mapping of the Sen2 gene [14] and was developed by crossing potato clones DG 97-264 (seed parent, P1) and DG 97-1805 (pollen parent, P2). Both parental clones were bred in IHAR-PIB Młochów Research Center, Poland, and both were complex interspecific Solanum hybrids. Species compositions of these clones were described by Plich and coworkers (2018). Parental clones differ in terms of tendency for tuber greening: the clone DG 97-264 has a strong tendency for greening while clone DG 97-1805 is more resistant (Table 1, Fig. 1). Progeny clones, parents and the standard potato cultivars Bintje, Evora, Jelly, Snowden and a diploid potato clone DG 97-2174 were field propagated to provide tubers for phenotypic tests.
Assessments of tendency for tuber greening
The progeny clones, along with parental clones and standards, were evaluated across three years 2015, 2016 and 2018. Tubers were harvested at the end of August and stored in the regular potato storage house at 4–8 °C for about 5 months. At the turn of January and February tubers were transferred to greenhouse and placed on tables lined with white paper. Tubers were exposed to natural light for two weeks. After this period they were visually evaluated in terms of two elements of tuber greening according to method described by Jakuczun [15]. The tuber external greening (hereinafter called tuber greening) was evaluated in 0–5 scale, where 0 = lack of greening and 5 = very intensive greening. Internal depth of greening (hereinafter called greening depth) was evaluated in tubers cut longitudinally into halves in 0–5 scale, where 0 = lack of greening, 1 = greening just below the skin, 2 = greening up to 2 mm, 3 = up to 5 mm, 4 = up to 10 mm, 5 = more than 10 mm deep. Each year 6–12 tubers per clone/cultivar were evaluated (3–6 tubers in two replications).
Statistical analyses
To assess reproducibility of tuber greening assessments between the individual years the Pearson’s correlation coefficients were calculated. To assess the influence of genotype, year, and their interactions on the tuber greening, the analysis of variance (ANOVA) was applied. Due to lack of some results of assessments of particular progeny clones from individual years, ANOVA was performed for results of evaluation of 124 individuals from SEN 12–01. Based on results of ANOVA a broad-sense heritability coefficient Hb was also calculated according to formula of Domański et al. [16]: Hb = σ2g/(σ2g + σ2ge + σ2e, σ2g = (M1 – M2)/L; σ2ge = M2—σe2, where M1 = mean square of effect of genotype, M2 = mean square of effect of genotype × year interaction, L = number of years, σ2e = mean square of error. Statistical analyses were performed with the use of MS Excel and STATISTICA 12.
Genetic mapping and QTL analysis
DNA isolation and DArTseq™ genotyping were performed as described by Plich et al. [14]. Total genomic DNA was extracted GenElute Plant Genomic DNA Miniprep Kit (Sigma-Aldrich, Germany) according to the supplier’s protocol. All progeny clones from the mapping population, along with the parental clones DG 97-264 and DG 97-1085, were genotyped by a genotyping-by-sequencing (GBS) approach (DArTseq™) at Diversity Arrays Technology Pty. Ltd. (Canberra, Australia) (https://www.diversityarrays.com/dart-application-dartseq). In our study only markers of PAV type (presence and absence variant), scored as 0 or 1, were used for map construction. Genotyping yielded the total amount of 53 731 DArTseq™ markers. The 4 704 DArTseq™ markers that were segregating and had missing data for less than 5% of individuals were used for grouping and genetic maps calculation in JoinMap® 4.1 (Van Ooijen [17]). Markers that were identical in terms of segregation or with similarity to each other ≥ 0.96 were excluded. The grouping settings were following: CP (cross pollination/outbreeder full-sib family) population type, independence LOD (significance cut-off, LOD score > 3) as a grouping parameter. Quality control and chromosome assignment of 12 linkage groups were carried out based on BLAST search of DArTseq™ sequences against a reference DM1-3 potato genome sequences (PGSC DM1-3 v 4.03). The markers mapped to chromosomes other than expected from the reference DM1-3 potato genome (PGSC DM1-3 v 4.03) were also excluded. Linkage maps were calculated using a regression method (linkage significance cut-off, LOD score > 3) and the Haldane’s mapping function for the calculation of map distances. QTL analysis was performed as described before [18, 19] using interval mapping with MapQTL®6 software [20]. QTLs were detected using an LOD threshold ≥ 3.0, estimated from cumulative distribution function of the maximum LOD on a chromosome for QTL analysis based on four QTL genotypes [21]. Average chromosome length in map SEN 12–01 was 81.01 cM, so the threshold LOD (α = 0.05) was between 2.9 (for chromosome length of 50 cM) and 3.2 (100 cM).
Results
The parents, 176 individuals of the mapping population SEN 12-01 and the five standards were evaluated for the two parameters of tuber greening in 2015, 2016 and 2018 (Table 1). The pollen parent DG 97-1805, with the mean tuber greening score 1.8 and tuber greening depth 0.4, showed significantly lower (1.8 times) tendency for tuber greening and tuber greening depth (3.2 times) than the seed parent DG 97-264 as indicated by a Duncan’s test. The parent DG 97-1805 did not differ significantly in tuber greening from the non-greening diploid standard, clone DG 97-2174, but showed significantly smaller greening depth that the standard. The greening parent DG 97-264 showed lower tuber greening (3.2) than all the standard tetraploid cultivars but the same level of tuber greening depth (1.3) as the least-greening cultivar Bintje. Standard cultivars Evora, Jelly and Snowden showed significantly higher tendency to tuber greening as well as tuber greening depth compared to both parental potato clones (Table 1, Fig. 1). Cultivar Snowden showed the most intensive tuber greening of all tested materials although some of the SEN 12-01 progeny clones exceeded it in tuber greening depth (Table 1, Fig. 1).
The distributions of the mean scores of greening parameters in the mapping population are shown in Fig. 2. The distributions were normal according to the Kolmogorov–Smirnov test but not according to the Shapiro–Wilk test. As these were distributions of mean values, the central limit theorem applies, further supporting their normality. The ranges of the mapping population individuals’ mean scores were similar for both greening parameters, and with the upper limit 3.5 they did not cover the full 0–5 scales (Table 1). However, the population mean and median were lower for the tuber greening depth (Table 1) which is also visible as a shift of tuber greening depth distribution towards lower values in Fig. 2. The scoring of the progeny individuals was more replicable between the three seasons of assessment for tuber greening depth than for tuber greening as shown by higher Pearson’s correlation coefficients (Table 1). Mean (2015, 2016, 2018) tuber greening and tuber greening depth in the population SEN 12-01 were strongly correlated with each other with Pearson’s correlation coefficient r = 0.739, p < 0.000.
Phenotypic distribution of mean (2015, 2016, 2018) tuber greening (a) and tuber greening depth (b) assessed in 0–5 scales among 176 individuals of the mapping population SEN 12-01. Score 0 = lack of tuber greening while score 5 = very intensive greening (for tuber greening) or greening deeper that 10 mm (for tuber greening depth). Scores obtained by the parental clones are marked: P1—DG 97-264, P2—DG 97-1805. Results of normality tests are shown above the charts; fitness to the normal curve is indicated by a line
Analysis of variance performed on the results of the tuber greening assessments in the population SEN 12-01 demonstrated significant effects of plant genotype, year of testing and genotype × year interaction on both tuber greening parameters (Table 2). While plant genotype explained the most of variance observed in tuber greening depth (45.71%), year of testing had the strongest effect on the tuber greening assessment (36.96%), and the interaction between those two factors contributed to the results of scoring both tuber greening parameters on similar level (16–17% of the variance explained; Table 2). The broad-sense heritability coefficient (Hb) for tuber greening was 0.464 and for tuber greening depth: Hb = 0.602.
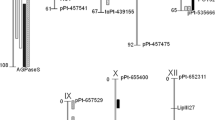
A genetic map common for both parents consisted of 1151 DArTseq™ markers and its total length was 973 cM. An average number of markers per chromosome was 95.9 and varied between 52 on chromosome XI and 175 on chromosome I (Fig. 3). Average chromosome length was 81.1 cM and it ranged from 53.5 cM (chromosome X) to 105.5 cM (chromosome I) (Fig. 3). A complete genetic map SEN 12-01 is shown in Supplementary Material 1 along with its alignment to the physical map of the reference DM1-3 potato genome (PGSC DM1-3 v 4.03). Almost half of the DArTseq™ marker sequences (489 of 1151 markers) were located in the reference DM1-3 potato genome and for 88.4% of those markers, the marker order on genetic map SEN 12-01 and on physical reference map was the same (Supplementary Material 1).
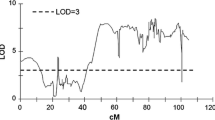
Interval QTL mapping was performed on three data sets of mean (2 replications × 3–6 tubers) scores from each year 2015, 2016 and 2018 separately and also on the mean dataset (mean score from 3 years of testing) for each greening parameter (Supplementary Materials 2 and 3). QTLs detected in mean datasets that were reproducibly detected also in all three data sets from particular years of testing are shown in Fig. 4 and Table 3.
LOD charts of QTL detected for 3-year mean (2015, 2016, 2018) tuber greening and tuber greening depth in the diploid potato mapping population SEN 12-01. Only QTL reproducibly detected in three years and with LOD > 3.0 are presented. y-axis: LOD, x-axis: chromosome genetic map (cM), threshold LOD = 3 marked by a line parallel to the x-axis
The two most important QTLs at LOD = 3 covered large regions of chromosomes VII and X and had the strongest effect on both greening parameters in all data sets. The strongest effect of the QTL on chromosome X on tuber greening was detectable at LOD ≥ 10 in the narrow interval between 21.2 and 24.6 cM. The strongest effect of that QTL on tuber greening depth was detected at LOD ≥ 7 in the interval 20.2–26.1 cM (Fig. 4). The LOD charts of those two QTLs overlapped for the two parameters with the peaks at very close positions on chromosome X at 23.2 cM. On chromosome VII, the peak for the mean tuber greening was at 31.3 cM while for the mean tuber greening depth it was at 28.3 cM (Fig. 4). Up to 16.9% of variance observed in the mean tuber greening (LOD = 7.1) could be ascribed to the QTL on chromosome VII and 23.4% (LOD = 10.17) to the QTL on chromosome X. The QTL on chromosome X had also a stronger effect on the mean tuber greening depth explaining up to 17.7% of variance (LOD = 7.45) in this trait, while the effect of the QTL on chromosome VII could be estimated at 13.1% of the explained variance (LOD = 5.38; Table 3). In the datasets from the particular years, the effect of QTL on chromosome X on the tuber greening varied (2015:17.1%, 2016: 11.1%, 2018: 26.3% of the explained variance) and the peak of the QTL shifted between 33.1 cM in 2015, 24.6 cM in 2016 and 26.3 cM in 2018. The effect of the QTL on chromosome X on the tuber greening depth changed between 12.6% in 2015, 14.9% in 2016 and 18.7% of variance explained in 2018 and its peak shifted: 22.2 cM in 2015, 23.2 cM in 2016 and 29.8 cM in 2018 (Supplementary Material 2). The effect of QTL on chromosome VII on the tuber greening varied (2015:15.9%, 2016: 12.6%, 2018: 21.7% of the explained variance) and the peak of the QTL shifted between 43.1 cM in 2015, 32.5 cM in 2016 and 28.3 cM in 2018. Similarly, the effect of the QTL on chromosome VII on the tuber greening depth changed between 12.4% in 2015, 12.6% in 2016 and 14.4% of variance explained in 2018 and its peak shifted: 39.1 cM in 2015, 8.4 cM in 2016 and 28.3 cM in 2018 (Supplementary Material 2).
For tuber greening the two QTLs on chromosomes VII and X were the only ones that were significant in all 3 years of phenotypic evaluation. Additional QTLs for this trait were also detected on chromosomes I (significant in 2015, 2018 and in the mean dataset explaining up to 10.7% of the variance), III (significant in 2015 and in the mean dataset explaining up to 8.6% of the variance), IV (significant only in 2016 and explaining up to 10% of the variance), VIII (significant in 2015, 2018 and in the mean dataset explaining up to 8.4% of the variance), IX (significant in 2015, 2018 and in the mean dataset explaining up to 10.7% of the variance), XI (significant in 2016 and in the mean dataset explaining up to 8.6% of the variance) and XII (significant in 2016 and in the mean dataset explaining up to 11.0% of the variance) (Supplementary Material 2).
The tuber greening depth was more stable across the year data sets and, apart from the QTLs on chromosome VII and X, three more QTLs were detected in all data sets: QTL on chromosomes I (up to 13.1% of variance explained in mean data set), III (up to 12.0% of variance explained in mean data set) and VIII (up to 13.6% of variance explained in mean data set) (Fig. 4, Table 3).
Additional QTLs for the tuber greening depth were detected in particular data sets only, on chromosomes: II (significant only in 2016 and explaining up to 8.3% of the variance), IV (significant in 2016 and in the mean dataset explaining up to 8.4% of the variance), IX (significant only in 2015 and explaining up to 14.4% of the variance) and XII (significant in 2016 and in the mean dataset explaining up to 8.6% of the variance) (Supplementary Material 2).
For tuber greening, no QTL was detected on chromosomes II, V and VI, and for tuber greening depth, no QTL was detected on chromosomes V, VI and XI in any of the datasets.
Discussion
Our work is the first analysis of QTLs for tuber greening resistance in potato, which is a step towards understanding of the genetic basis of this trait and towards efficient breeding of potatoes resistant to tuber greening. Theoretically, tuber greening may be regulated at numerous stages of light perception, signal transduction, amyloplast transformation and chlorophyll pigments synthesis and degradation. The intensity of tuber greening may be affected by the allelic diversity of a large number of genes involved in those processes but also by the factors like skin colour, thickness and texture, tuber flesh colour and density, as well as many environmental factors [4]. Our data only partially confirmed previous reports and our main hypothesis on polygenic inheritance of tuber greening [7,8,9,10,11]. The described landscape of the potato genome regions significant for tuber greening resistance is an indication that in spite of the large number of the genes potentially important for the trait, some factors located on the chromosomes VII and X play key roles and determine big parts of the observed tuber greening variation under natural light exposure in the greenhouse. We suppose that experiments under controlled light conditions might significantly reduce both influence of the year of testing on the traits and an error of the experiment.
The external tuber greening and tuber greening depth were correlated and the most important QTLs, that affected them, overlapped. Additional, minor QTLs were year- and/or trait-specific, which is in agreement with the results obtained by Jakuczun and Zimnoch-Guzowska [11], who postulated that the external tuber greening and its depth may be partially independently inherited. In contrast, an earlier study which has dissected tuber greening into three components (external colour, internal colour and its depth), has suggested that they are inherited independently and all three of them should be considered in breeding programs aiming at tuber greening resistance [10]. These differences may be related to the use of genetically different materials in listed studies and to the differences in tuber skin and flesh colour, thickness, texture, etc. Our material was derived from complex interspecific hybrids having S. tuberosum, S. chacoense, S. yungasense, S. gourlayi, S. acaule, S. demissum, S. verrucosum, S. microdontum and S. phureja in their pedigrees [14], and had rather uniform skin and flesh without noticeable anthocyanin pigmentations.
The interspecific hybrid nature of our material might have affected also the comparison of the SEN 12-01 genetic map with the reference genome S. phureja DM1-3 (Supplementary Material 1). The SEN 12-01 genetic map is the first genetic map of the full potato genome constructed with DArTseq™ markers. Its length and density are comparable with earlier DArT maps for which the markers were generated by hybridization methods [18, 19]. A main advantage of DArTseq™ markers was the possibility of comparison of the marker order on our genetic map to the physical map of the reference genome, by a BLAST search of the obtained sequences. That resulted in locating 489 of 1151 markers in the reference potato genome DM1-3 and for 88.4% of those markers the marker order on both maps was the same. The fact that majority of DArTseq™ (662) sequences could not be unambiguously identified in the reference genome and that there were 11.6% of markers located to the discrepant positions, we explain rather by genuine differences between genomic sequences of our material and DM1-3, although some mapping errors cannot be excluded.
When we compared the location of QTLs for tuber greening parameters with the location of QTLs for glycoalkaloid content identified in other studies, in general we confirmed the independent inheritance of the two traits. The accumulation of both glycoalkaloids and chlorophylls in tubers is stimulated by light but it occurs through different metabolic pathways [4]. Still some light perception elements and regulatory factors could be common for both processes. There are only a few works on inheritance and QTLs mapping of factors affecting glycoalkaloid content in potato tubers, whereas the leaf content of glycoalkaloids has been investigated more frequently. The earliest study of glycoalkaloid content in potato tubers has demonstrated that it is a trait affected by a single locus, modulated by two interacting loci of unknown genome location [22]. Sørensen et al. [23] have mapped a major QTL associated with glycoalkaloid content, with and without an additional post-harvest light exposure, to chromosome I using two BC1 populations of S. tuberosum and S. sparsipilum. That result was confirmed using the same material and new technology, next generation sequencing bulk segregant analysis, and the QTL was located on the physical map of the potato genome to the interval between 63.1 and 73.5 Mb of chromosome I [24]. In this work two more QTLs for glycoalkaloid content in potato tubers have been identified with less confidence on chromosome VI (13.3–15.4 Mb) and VIII (4.8–8.0 Mb). While none of our two major QTLs on chromosomes VII and X overlapped with QTLs for tuber glycoalkaloid content, some partial overlaps could be noted on chromosomes I and VIII, where we detected weak QTLs for tuber greening and for tuber greening depth (Supplementary Materials 1 and 2). Prominent QTLs for various foliar glycoalkaloids contents in potato have been identified on chromosome I, including QTL for solanidine [25], leptine [26] and leptinine [27] content. Due to the use of different marker systems in those studies it is not possible to compare directly the locations of QTLs for foliar and tuber glycoalkaloid content. However, the DNA sequence of the most significant marker TG70 [25] from a tomato clone can be found in the GABI Primary Database [28], and it can be located in the potato reference genome (DM v4.04) at 62 Mb which is close to the QTL for tuber glycoalkaloid content detected previously [23, 24] but also to a minor QTL for tuber greening detected in our study (Supplementary Materials 1 and 2).
The main conclusion from our study is that factors located on the chromosomes VII and X play key roles and determine big parts of the observed tuber greening variation. Markers tagging those regions could be useful for selection of potato clones less susceptible to greening.
References
FAO (2019). https://www.fao.org/faostat/en/#data/QC. Accessed 25 Apr 2019
Chang HY (2013) Light-induced glycoalkaloid and chlorophyll synthesis in potato tubers: Cultivar differences and strategies for mitigation. MS thesis, University of California
Van der Merwe L (2016) Most important reasons for downgradings on markets for certain regions—2015/2016. CHIPS November/December 2016: 42–48. Potatoes South Africa. https://www.potatoes.co.za/media-events/chips/chips-november/december-2016.aspx. Accessed 8 Feb 2017
Tanios S, Eyles A, Tegg R, Wilson C (2018) Potato tuber greening: a review of predisposing factors, management and future challenges. Am J Potato Res 95:248–257
Percival G (1999) Light-induced glycoalkaloid accumulation of potato tubers (Solanum tuberosum L.). J Sci Food Agric 79:1305–1310
Grunenfelder LA, Knowles LO, Hiller LK, Knowles NR (2006) Glycoalkaloid development during greening of fresh market potatoes (Solanum tuberosum L.). J Agric Food Chem 54:5847–5854
Akeley RV, Houghland GVL, Schark AE (1962) Genetic differences in potato tuber greening. Am J Potato Res 39:409–417
Brown E, Riley W (1976) Greening of potato tubers: varietal response to controlled exposure to light. J Natl Inst Agric Bot 14:70–76
Parfitt DE, Peloquin SJ (1981) The genetic basis for tuber greening in 24-chromosome potatoes. Am Potato J 58:299–304
Reeves AF (1988) Varietal differences in potato tuber greening. Am J Potato Res 65:651–658
Jakuczun H, Zimnoch-Guzowska E (2006) Inheritance of tuber greening under light exposure in diploid potatoes. Am J Potato Res 83:211–221
Bamberg J, Moehninsi M, Navarre R, Suriano J (2015) Variation for tuber greening in the diploid wild potato Solanum microdontum. Am J Potato Res 92:435–443
Novy R, Schneider B, Fristad M, Schroeder L, Stark J, Kuhl J (2018) Introgression of resistance to tuber greening from Solanum microdontum into cultivated potato. Am J Potato Res 95:208–229
Plich J, Przetakiewicz J, Śliwka J, Flis B, Wasilewicz-Flis I, Zimnoch-Guzowska E (2018) Novel gene Sen2 conferring broad-spectrum resistance to Synchytrium endobioticum mapped to potato chromosome XI. Theor Appl Genet 131:2321–2331. https://doi.org/10.1007/s00122-018-3154-y
Jakuczun H (2001) Assessment of the tendency to greening in potato tubers exposed to light. IHAR Monografie i Rozprawy Naukowe, 10 A. Plant Breeding and Acclimatization Institute, Radzików, pp 109–110
Domański L, Michalak K, Zimnoch-Guzowska E (2007) Variation of blackspot susceptibility of the selected potato cultivars. Biuletyn IHAR 246:145–149
Van Oojien JW (2006) JoinMap®4, Software for the calculation of genetic linkage maps in experimental populations. Kyazma B.V., Wageningen
Hara-Skrzypiec A, Śliwka J, Jakuczun H, Zimnoch-Guzowska E (2018) Quantitative trait loci for tuber blackspot bruise and enzymatic discoloration susceptibility in diploid potato. Mol Genet Genomics 293:331–342
Śliwka J, Sołtys-Kalina D, Szajko K, Wasilewicz-Flis I, Strzelczyk-Żyta D, Zimnoch-Guzowska E, Jakuczun H, Marczewski W (2016) Mapping of quantitative trait loci for tuber starch and leaf sucrose contents in diploid potato. Theor Appl Genet 121:131–140
Van Ooijen JW (2009) MapQTL ® 6, Software for mapping of quantitative trait loci in experimental populations of diploid species. Kyazma BV, Wageningen
Van Ooijen JW (1999) LOD significance thresholds for QTL analysis in experimental populations of diploid species. Heredity 83:613–624
van Dam J, Levin I, Struik PC, Levy D (2003) Identification of epistatic interaction affecting glycoalkaloid content in tubers of tetraploid potato (Solanum tuberosum L.). Euphytica 134:353–360
Sørensen KK, Kirk HG, Olsson K, Labouriau R, Christiansen J (2008) A major QTL and an SSR marker associated with glycoalkaloid content in potato tubers from Solanum tuberosum × S. sparsipilum located on chromosome I. Theor Appl Genet 117:1–9
Kaminski KP, Kørup K, Andersen MN, Sønderkær M, Andersen MS, Kirk HG, Nielsen KL (2016) Next generation sequencing bulk segregant analysis of potato support that differential flux into the cholesterol and stigmasterol metabolite pools is important for steroidal glycoalkaloid content. Potato Res 59:81–97. https://doi.org/10.1007/s11540-015-9314-4
Yencho GC, Kowalski SP, Kobayashi RS, Sinden SL, Bonierbale MW, Deahl KL (1998) QTL mapping of foliar glycoalkaloid aglycones in Solanum tuberosum × S. berthaultii potato progenies: quantitative variation and plant secondary metabolism. Theor Appl Genet 97:563–574
Ronning CM, Sanford LL, Kobayashi RS, Kowalski SP (1998) Foliar leptine production in segregating F1, Inter-F1, and backcross families of Solanum chacoense bitter. Am J Potato Res 75:137–143
Hutvágner G, Bánfalvi Z, Milánkovics I, Silhavy D, Polgár Z, Horváth S, Wolters P, Nap J-P (2001) Molecular markers associated with leptinine production are located on chromosome 1 in Solanum chacoense. Theor Appl Genet 102:1065–1071
GABI Primary Database https://www.gabipd.org/. Accessed 30 July 2019
Acknowledgements
This study was founded by National Science Center in Poland (Grant 2013/11/B/NZ9/01959). It was also supported by a statutory dotation 1-3-00-3-05 (JŚ) and 1-3-00-3-03 (JP, BT and EZG) from the Polish Ministry of Science and Higher Education in 2018 and 2019.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Plich, J., Zimnoch-Guzowska, E., Tatarowska, B. et al. Quantitative trait loci analysis of potato tuber greening. Mol Biol Rep 47, 1713–1722 (2020). https://doi.org/10.1007/s11033-020-05263-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05263-3