Abstract
In horses, the identification of the genetic background of phenotypic variation, especially with regard to performance characteristics and predisposition to effort, has been extensively studied. As α-actinin-3 function is related to the regulation of muscle contraction and cell metabolism, the ACTN3 gene is considered one of the main genetic factors determining muscle strength. The aim of the present study was to assess the genotype distribution of two SNP variants within the equine ACTN3 gene (g.1104G > A and c.2334C > T) across different utility types and horse breeds. The analyses were performed on five breeds representing horses of different types, origins and utilities according to performance (in total 877 horses): primitive (Polish koniks; Hucul horses), draught (Polish heavy draught) and light (Thoroughbred and Arabian horses). Two polymorphisms within the ACTN3 gene locus were genotyped and genotype and allele frequency were compared across populations in order to verify if the identified differences contribute to the phenotypic variation observed in horse breeds. The present study allowed confirmation of the significant differences in genotype distribution of g.1104G > A localized in the promoter region between native breeds and racehorse breeds such as Thoroughbreds and Arabians. The allele/genotype variations between primitive and light breeds confirmed that the analysed variant was under selection pressure and can be correlated with racing ability. Moreover, the significant differences for the c.2334C > T genotype frequency between Arabian horses and other breeds indicate its relationship with endurance and athletic performance. The predominance of the T allele (85%) in Arabians suggests that the T variant was favoured during selection focused on improving stamina and could be one of the genetic factors determining endurance ability. Further research is needed to confirm the association of both polymorphisms with exact racing and/or riding results.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Alpha-actinins are 100-kDa proteins which belong to the superfamily of cross-linking actin filament proteins. Among them, two muscular-specific isoforms are found, namely, α-actinin-2 and α-actinin-3, as well as 2 non-muscular isoforms, namely, α-actinin-1 and α-actinin-4 [2, 3]. Muscle proteins constitute a part of the contractile mechanism by anchoring actin fibres on the Z line in striated muscle and in dense bodies in smooth muscle cells [13]. In humans, α-actinin-2 occurs in all types of skeletal muscle fibres (I, IIA and IIB), whereas α-actinin-3 is present only in type II fibres, termed fast twitch [12]. The role of α-actinin-3 is crucial during muscle contraction because it is involved in anchoring fibre–actin filaments. In addition, α-actinin-3, due to its spatial position in the cell near the enzyme glycogen phosphorylase, regulates its activity and influences glycogen metabolism in muscle tissue [1, 16].
In humans, the ACTN3 gene, which codes for α-actinin-3, is considered the main determining factor for the predisposition to short or long-term effort (sprint or endurance ability). In addition, polymorphisms within the ACTN3 gene are already used in commercial genetic tests to assess the performance phenotype of athletes and for the optimal selection of training due to individual predispositions [6]. The functional and structural analysis of the ACTN3 gene is also performed with regard to its association with horse performance characteristics. To date, several reports have focused on the identification of SNP variations in the equine actinin-3 gene in different breeds [10, 24, 26]. Moreover, in horses, the modification of ACTN3 transcript abundance under training conditions was confirmed [20, 21]. In studies conducted on purebred Arabian horses, a significant decrease in the level of ACTN3 gene expression in muscle tissue was detected during long-term flat race training using an RNA-seq approach [21] and accurate real-time PCR method [20]. The study of Quinlan et al. 92010) showed that alpha-actinin-3 is related to regulation of glycogen phosphorylase activity resulting in a metabolic shift promoting the increase in fatty acid use in ATP generation. Thus, it can be hypothesized that (decreased of ACTN3 expression level in muscle can be associated with metabolism regulation [20].
The above reports indicate an important role of α-actinin-3 in the process of adaptation to physical effort. Therefore, there is a need to conduct studies to identify polymorphisms in the ACTN3 gene in horses, which can be correlated with performance characteristics and could be used in the future as a genetic marker. The aim of present study was to assess the genotype distribution of two SNP variants across different utility types and horse breeds (g.1104G > A and c.2334C > T). The analysed polymorphisms were considered to be potentially related to equine ACTN3 gene transcription, and the differences in genotype/allele frequency between analysed breeds can contribute to the phenotypic variation observed in horse breeds.
Materials and methods
Animals
A total of 877 horses were included in the study. The analyses were performed on five breeds representing horses of different types, origins and utilities according to performance: primitive, draught and noble light. Samples from primitive included: Polish koniks (n = 182) and Hucul horses (n = 78). Heavy draught included: Polish heavy draught—Polish Coldblood Horse—PKZ (n = 76). Noble light breeds included: Arabian horses (n = 450) and Thoroughbred horses—TB (n = 91). All samples were previously collected and stored in the Biological Material Bank of the National Research Institute of Animal Production and in the Department of Horse Breeding, Institute of Animal Science, the University of Agriculture in Krakow. The whole blood or hair follicle samples were collected previously as part of other research projects.
Polish koniks
The analysed primitive horses belonging to the Polish koniks breed included representatives of 16 existing maternal and 6 paternal lines. The sampled horses were raised in privet breeders and were included in planned breeding or were representatives of a semi-feral population maintained at the primeval Białowieza Forest. The Polish konik is a native breed presenting behaviour adapted to local environmental conditions, as only these horses are maintained in all possible maintenance systems (herds in conditions close to natural, groups in stable-less systems and individuals in stables). Polish koniks are perfectly adapted to life in forest conditions due to a strong and stocky build, excellent healthiness, and a primitive dun coat and dorsal stripe [23]. They are also used in recreational riding.
Hutsul horses
The second primitive breed, the Hutsul horses, were sampled in State Stud. The first written reference to the supposed Hutsul horses comes from 1603 AD and was published by K. Drohostajski in “Hippika”. Hutsul horses were raised in the Eastern Carpathians in the Hutsul region and Bukovina, and in the upper reaches of Czeremosz, Prut, Putilla, Moldova and Suceava. The origin of Hutsul horses it is not clear. It is thought that they are descendants of various types of Tatar horses, Polish, Oriental, Turkish, tarpans, Przewalski horses, and Nrse horses. The breed was consolidated in the 19th century in the Eastern Hutsul region and the Eastern Carpathians under the influence of harsh climate, poor pasture and primitive living conditions. Hutsul is considered one of the oldest primitive breeds maintained in Poland and due to the excellent adaptation to mountain environment conditions are characterized by a strong and lean constitution, longevity, strength and disease resistance [15].
Polish heavy draught
The polish heavy draught horses represent the Sztumski type. Samples were collected from a State stud herd with breeding purposes to provide breeding material to improve locally maintained herds. In fact, the breeding of cold-blooded horses is focused on yielding individuals well suited for draught power and for meat production purposes [18]. The draught horses are heavy boned, with large musculature consisting of short and slow oxidative fibres [5].
Thoroughbred horses
The Thoroughbred horse is one of the most noble breeds in world. Raised more than 300 years ago in UK, the intensive selection focused on a race-athletic phenotype leading to a breed with spectacular achievements and racecourse performance. The samples in this study were collected from horses maintained in state stud with a breeding history since the second world war (IIWW). The individuals are descendants of 36 stallions and 59 dams.
Arabian horses
The analysed Arabian horses represent the population of Polish Arabian horses maintained in State Studs belonging to 16 maternal and 9 paternal lines with a confirmed pure breed of more than 200 years. Arabian horses compete in an equestrian sport, however, they dominate in endurance competitions. Furthermore, they are often used to enrich not only refinement in other horse breeds but also endurance abilities [30]. The selection in Arabians is focused on improving traits related to endurance and athleticism.
Whole blood or hair follicles were collected from the horses. The protocol was approved by the Animal Care and Use Committee of the Institute of Pharmacology, Polish Academy of Sciences in Kraków (no.1173/2015).
SNPs identification
For all horses, two polymorphisms were selected for further evaluation of association with performance characteristics. A previous studies allowed the identification of two Single Nucleotide Polymorphisms (SNPs)—one in the promoter region (g.1104G > A; rs1144978872) [24] and one in the 19th exon located near a splice region in the junction between the 18th and 19th exons (c.2334C > T) (c.2334C > T; rs1141508235) [20], which were detected in Arabian horse population. Both SNPs can potentially affect the transcription process of the ACTN3 gene.
The polymorphisms were genotyped using the PCR–RFLP method according to the procedure presented in Table 1. The PCR product was obtained using Phanta Master Mix and High-Fidelity PCR (Vazyme Biotech Co., Ltd) according to the manufacturer’s protocol with an annealing temperature at 57 °C. After amplicon digestion, the PCR products were separated on 3% and 4% agarose gels (for g.1104G > A and c.2334C > T; respectively).
The Hardy–Weinberg equilibrium was calculated using the Court Laboratory HW calculator (Court’s 2005–2008 online calculator; [4]). The differences in genotype distribution between breeds were estimated using the Chi square test according to (Preacher [14]).
Results and discussion
Genotype and allele distribution of g.1104G > A SNP
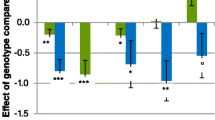
The genotyping of the g.1104G > A polymorphism in a total of 877 horses showed significant differences in genotype and allele frequencies across the analysed breeds. The three native and coldblooded breeds were characterized by the predominance of the AA genotype and the low number of GG horses (Table 2; Fig. 1). The lowest number included animals with the GG genotype in both native Polish koniks and Hucul horses (1% and 1.9% respectively). The opposite genotype distribution was detected in noble breeds: Arabian and Thoroughbred had GG horses accounting for 47.7% in both analysed populations (Fig. 1). In TB horses, no AA horses were identified. Furthermore, both Arabian and Thoroughbred populations were not consistent with HWE equilibrium (Table 2). The analysis of allele and genotype distribution confirmed the significant (p < 0.0001) differences between light breeds and other breeds. Moreover, both light breeds also differed between each other (genotype distribution—p < 0.035; alleles frequency p < 0.0001) (Fig. 2; Table 3).
Genotype and allele distribution of c.2334C > T SNP
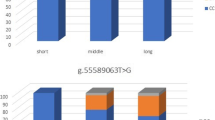
The analysis of the c.2334C > T allele and genotype distribution showed less variation between the investigated breeds than the g.1104G > A polymorphism. A similar genotype frequency was identified for Polish koniks and Thoroughbreds. For Hutsul horses, three genotypes were also identified; the most abundant were heterozygotes. In the Polish coldblood population, only CC and TC horses were identified (Fig. 3; Table 2). For most breeds, a higher frequency was detected for the CC genotype (form 86.6% for PKZ to 42.6% for Hutsul horse). The opposite genotype frequency was observed in the Arabian horse population, where CC horses accounted for 2% of all horses (Fig. 3). This was confirmed by differences in C–T allele distribution across analysed breeds and the Arabian horse population (< 0.0001) (Fig. 4; Table 4).
Discussion
The association between muscle histological structure (the percentage and diameter of different fibre types) and predisposition to a specific type of effort (sprint or endurance) has been established [27, 31]. The percentage of individual muscle fibres and their diameter depend on many factors, including environmental factors (nutrition or breeding) as well as genetic background [28]. Studies conducted in humans have shown the relationship between the proportion of specific types of muscle fibres and the type of physical exercise performed. In people practicing strength discipline or runners, fast-twitch fibres predominate, while in endurance sports, slow-twitch fibres are more abundant [11, 27, 31]. The α-actinin-3 is fibre type specific and presents only in type II fast twitch fibres [12]. Moreover, a significant correlation was found between the gene encoding α-actinin-3 with the percentage of particular types of muscle fibres [25]. Due to α-actinin-3 localization and function, the fast fibre-specific gene ACTN3 has been proposed as one of the critical genetic factors determining skeletal muscle function and metabolism [1].
In humans, the role of the ACTN3 gene has been thoroughly studied, making it possible to determine its effect on the phenotype associated with physical exercise. The mutation present in the ACTN3 gene, located in position R577X (rs1815739), has a significant impact on the predisposition of the unit for short and fast or long-distance runs. This mutation occurs in a large part of the population, in which it causes a deficiency of the α-actinin-3 protein. It has been established that the R577X polymorphism directly leads to a change in the features of muscle fibres. The R577X mutation is located at position 577 of the gene encoding α-actinin-3 and results in the conversion of cytosine to thymine, C > T. As a consequence, the amino acids sense change and instead of the appearance of the next amino acid–arginine, a premature STOP codon is created. Studies conducted in humans have shown that α-actinin-3 is absent in 18% of healthy, light-skinned individuals due to the presence of the homozygous TT genotype [29]. The association of ACTN3 R5777X polymorphism with power and sprint performance has been established by numerous researches [6, 8, 9, 17]. On the other hand, the relation of R5777X to endurance performance [17, 19, 22] and its genetic model of inheritance (dominant, additive, recessive) [7] still have not been completely known.
The huge potential of using the ACTN3 gene as a genetic marker in athletes led to its analysis in other species, especially in horses, which are evolutionarily adapted to effort and are a perfect model for adapting the body to exercise and training. The sequence of the horse ACTN3 gene encoding alpha-actinin-3 was previously studied by Mata et al. [10] and Thomas et al. [24]. Based on sequencing of the ACTN3 gene performed in 15 horses, Thomas et al. [24] identified 34 SNPs. Most of them (28 SNPs) were located in the non-coding region, and the remaining 6 polymorphisms in exons were discovered as silent changes. The authors also identified several new DNA sequence variations, of which, g.1104G > A was proposed as the most interesting, with a potentially large effect on the function of the alpha-actinin-3 protein due to its location in the 5′UTR region and with a different frequency of this change in the breeds of horses with different predispositions to effort. Thomas et al. [24] hypothesized that the 5′UTR variant polymorphism may affect horse racing characteristics by modifying the level of ACTN3 gene expression. This hypothesis was supported by Ropka-Molik et al. [20], which showed that flat-racing training in Arabian horses significantly modified ACTN3 gene expression regardless of the analysed splice isoforms. Moreover, the authors indicated a possible effect of the g.1104G > A polymorphism on ACTN3 gene expression in the gluteus medius muscle in horses after an intense gallop phase [20].
In 2014, Thomas et al. presented the genotype and allele frequency of the ACTN3 g.1104G > A SNP in sprint (Thoroughbred), endurance (Arabian), pace (Starnardbred) and strength (Clydesdale, Shire) types horses. The research confirmed the differences in both genotype and allele distributions between analysed breeds. The warmblood horses such as Thoroughbred and Arabian horses have a higher percentage of the G allele - 83% and 62%, respectively, while Standardbred and Clydesdale/Shire horses (coldblooded breeds) have only 31% and 23% of the G allele, respectively [24]. The results obtained in the present study confirmed the low frequency of the G allele in the native breed (Polish koniks and Hutsul) and Polish heavy draught (20%, 17% and 36%, respectively), 60% in Thoroughbreds and 74% in Arabian horses. Similar to the report by Thomas et al. [24], only the GG and AG genotypes were identified in TB horses. The opposite genotype frequency was detected for the muscular and powerful horse breeds Clydesdale and Shire, where no GG horses were found and the majority of the analysed group were AA animals (86%). Interestingly, in the present study the predominance of AA horses was identified in both native breeds, Polish konik (65%) and Hucul horses (69%), while in the Polish heavy draught population AA accounted for only 42%. The analysis of all five horse populations showed a significant increase in the G allele frequency from the native breed throughout coldblood to sprint and endurance types (from 17% in Hucul to 74% in Arabian). Such an identified trend indicated an association of allele A with the primitive type of horse breed and strength. On the other hand, the significant predominance of the G allele in warmbloods, especially Arabian horses, directed it to racehorse breeding.
With regard to the second analysed SNP c.2334C > T localized in the 19th exon, highly significant differences were observed between Arabian horses and other breeds (p < 0.0001). In Arabian horses, the frequency of the T allele was 85%, while in sprint type TB horses it was only 14% and only 7% in Polish heavy draught horses. In the coldblood population, no TT horses were identified. Surprisingly, no significant differences in allele and genotype distributions were obtained between Thoroughbred and other breeds, where the CC horses predominated. The c.2334C > T polymorphism is a synonymous variant with a potential low effect on protein structure or gene function. However, the analysed SNP is localized in the vicinity of the splice region at the exon junction and can be related to modification of precursor messenger RNA into mature messenger RNA in the splicing process. Arabian horses are characterized by unusual strength, stamina and dominance in endurance riding. The selection in this breed is focused on improving traits related to endurance and athleticism. Therefore, the distribution of the c.2334C > T genotype strongly indicates that the T allele can be essential for endurance ability, while the lack of a difference between sprint type horses and native and coldblooded breeds showed that the investigated SNP is not associated with sprint performance traits.
Both analysed polymorphisms (g.1104G > A; c.2334C > T) were also identified in Chinese native Yili Horses [26]. In this analysed breed, the authors showed a similar genotype distribution of the g.1104G > A polymorphism as found in our Arabian horse population (for Yili horses AG–44.74 and GG–42.11% and for Arabian horses AG–53.1, GG–47.7%). Furthermore, for c.2334C > T SNP genotype frequency (CC–86.84%; CT–10.53%; TT–2.63%), the Yili Horse population was the most similar to Polish koniks and completely opposite of Arabian horses. Wang et al. [26] focused on another SNP missense variant, g.9059T > G, identified in this breed and proposed it as a clausal mutation affecting athletic performance (TT–94.74% and TG–5.26%). The authors showed that g.9059T > G changed the secondary structure of α-actinin-3, and proposed this SNP as a candidate for breeding improvement towards racing traits.
Conclusion
The present study allowed a comparison of the allele and genotype frequencies of ACTN3 polymorphisms across horses representing different utility types and breeds. The results confirmed the significant differences in genotype distribution of g.1104G > A localized in the promoter region between native breeds and racehorse breeds such as TB and Arabians. Such discrepancies indicated that g.1104G > A was under selection pressure since horse domestication is focused on improving desired features. The allele/genotype variations between primitive and light breeds show that the analysed variant is related to racing ability. Furthermore, highly significant differences in the genotype frequency of c.2334C > T between Arabian horses and other breeds can indicate its relationship with endurance and athletic performance. The predominance of the T allele (85%) in Arabians could suggest that the T variant was favoured during selection focused on improving stamina and could be one of the genetic factors determining endurance ability. Further research is needed to confirm the association of both polymorphisms with exact racing and/or riding results.
References
Berman Y, North KN (2010) A gene for speed: the emerging role of alpha-actinin-3 in muscle metabolism. Physiology 25:250–259
Beggs AH, Byers TJ, Knoll JH, Boyce FM, Bruns GA, Kunkel LM (1992) Cloning and characterization of two human skeletal muscle alpha-actinin genes located on chromosomes 1 and 11. J Biol Chem 267:9281–9288
Blanchard A, Ohanian V, Critchley D (1989) The structure and function of alpha-actinin. J Muscle Res Cell Motil 10:280–289
Court MH, Michael H (2012) Court’s (2005–2008) online calculator. (Tuft University). http://goo.gl/1UElb4. Accessed 23 Jun 2016
Dall’Olio S, Fontanesi L, Nanni Costa L, Tassinari M, Minieri L, Falaschini A (2010) Analysis of horse myostatin gene and identification of single nucleotide polymorphisms in breeds of different morphological types. J Biomed Biotechnol 2010:542945
Eynon N, Hanson ED, Lucia A, Houweling PJ, Garton F, North KN, Bishop DJ (2013) Genes for elite power and sprint performance: ACTN3 leads the way. Sports Med 43:803–817
Garton F, North KN (2016) The effect of heterozygosity for the ACTN3 null allele on human muscle performance. Med Sci Sports Exerc 48:509–520
Garton FC, Houweling PJ, Vukcevic D, Meehan LR, Lee FXZ, Lek M, Roeszler KN, Hogarth MW, Tiong CF, Zannino D, Yang N, Leslie S, Gregorevic P, Head SI, Seto JT, North K (2018) The effect of ACTN3 gene doping on skeletal muscle performance. Am J Hum Genet 102:845–857
Kim H, Song KH, Kim CH (2014) The ACTN3 R577X variant in sprint and strength performance. J Exerc Nutr Biochem 18:347–353
Mata X, Vaiman A, Ducasse A, Diribarne M, Schibler L, Guérin G (2012) Genomic structure, polymorphism and expression of the horse alpha-actinin-3 gene. Gene 491:20–24
Mero A, Jaakkola L, Komi PV (1991) Relationships between muscle fibre characteristics and physical performance capacity in trained athletic boys. J Sports Sci 9:161–171
North KN, Beggs AH (1996) Deficiency of a skeletal muscle isoform α-actinin (α-actinin-3) in merosin-positive congenital muscular dystrophy. Neuromusc Disord 6:229–235
Otey CA, Carpen O (2004) Alpha-actinin revisited: a fresh look at an old player. Cell Motil Cytoskeleton 58:104–111
Preacher KJ (2001) Calculation for the Chi square test: an interactive calculation tool for Chi square tests of goodness of fit and independence [Computer software]. http://quantpsy.org
Purzyc H (2007) A general characteristic of Hucul horses. Acta Sci Pol 6(4):25–31
Quinlan KG, Seto JT, Turner N, Vandebrouck A, Floetenmeyer M, Macarthur DG, Raftery JM, Lek M, Yang N, Parton RG, Cooney GJ, North KN (2010) Alpha-actinin-3 deficiency results in reduced glycogen phosphorylase activity and altered calcium handling in skeletal muscle. Hum Mol Genet 19:1335–1346
Pickering C, Kiely J (2017) ACTN3: more than just a gene for speed. Front Physiol 8:1080
Polak G (2019) Genetic variability of coldblooded horses, participating in genetic resources conservation programs, using pedigree analysis. Ann Anim Sci 19(1):49–60
Rankinen T, Fuku N, Wolfarth B, Wang G, Sarzynski MA, Alexeev DG, Ahmetov II, Boulay MR, Cieszczyk P, Eynon N, Filipenko ML, Garton FC, Generozov EV, Govorun VM, Houweling PJ, Kawahara T, Kostryukova ES, Kulemin NA, Larin AK, Maciejewska-Karłowska A, Miyachi M, Muniesa CA, Murakami H, Ospanova EA, Padmanabhan S, Pavlenko AV, Pyankova ON, Santiago C, Sawczuk M, Scott RA, Uyba VV, Yvert T, Perusse L, Ghosh S, Rauramaa R, North KN, Lucia A, Pitsiladis Y, Bouchard C (2016) No evidence of a common DNA variant profile specific to world class endurance athletes. PLoS ONE 110:e0147330
Ropka-Molik K, Stefaniuk-Szmukier M, Musiał AD, Piórkowska K, Szmatoła T (2019) Sequence analysis and expression profiling of the equine ACTN3 gene during exercise in Arabian horses. Gene 685:149–155
Ropka-Molik K, Stefaniuk-Szmukier M, Żukowski K, Piórkowska K, Bugno-Poniewierska M (2017) Exercise-induced modification of the skeletal muscle transcriptome in Arabian horses. Physiol Genomics 49:318–326
Saunders CJ, September AV, Xenophontos SL, Cariolou MA, Anastassiades LC, Noakes TD, Collins M (2007) No association of the ACTN3 gene R577X polymorphism with endurance performance in Ironman Triathlons. Ann Hum Genet 71:777–781
Stefaniuk-Szmukier M, Ropka-Molik K, Piórkowska K, Szmatoła T, Długosz B, Pisarczyk W, Bugno-Poniewierska M (2017) Variation in TBX3 gene region in dun coat color Polish Konik horses. J Equine Vet Sci 49:60–62
Thomas KC, Hamiton NA, North KN, Houweling PJ (2014) Sequence analysis of the equine ACTN3 gene in Australian horse breeds. Gene 538:88–93
Vincent B, De Bock K, Ramaekers M, Van den Eede E, Van Leemputte M, Hespel P, Thomis MA (2007) ACTN3 (R577X) genotype is associated with fiber type distribution. Physiol Genomics 32:58–63
Wang J, Meng J, Wang X, Zeng Y, Li L, Xin Y, Yao X, Liu W (2018) Analysis of equine ACTN3 gene polymorphisms in Yili horses. J Equine Vet Sci 70:101–106
Wang YX, Zhang CL, Yu RT, Cho HK, Nelson MC, Bayuga-Ocampo CR, Ham J, Kang H, Evans RM (2004) Regulation of muscle fiber type and running endurance by PPARδ. PLoS Biol 2:e294
Wimmers K, Fiedler I, Hardge T, Murani E, Schellander K, Ponsuksili S (2006) QTL for microstructural and biophysical muscle properties and body composition in pigs. BMC Genet 9:7–15
Yang N, MacArthur DG, Gulbin JP, Hahn AG, Beggs AH, Easteal S, North K (2003) ACTN3 genotype is associated with human elite athletic performance. Am J Hum Genet 73(3):627–631
Zechner P, Sölkner J, Bodo I, Druml T, Baumung R, Achmann R, Marti E, Habe F, Brem G (2002) Analysis of diversity and population structure in the Lipizzan horse breed based on pedigree information. Livest Prod Sci 77(2–3):137–146
Zierath JR, Hawley JA (2004) Skeletal muscle fiber type: influence on contractile and metabolic properties. PLoS Biol 10:e348
Funding
The study was supported by the National Science Centre (Project No. 2014/15/D/NZ9/05256).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Musiał, A.D., Ropka-Molik, K., Piórkowska, K. et al. ACTN3 genotype distribution across horses representing different utility types and breeds. Mol Biol Rep 46, 5795–5803 (2019). https://doi.org/10.1007/s11033-019-05013-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-019-05013-0