Abstract
This is the first study which describes a unique procedure of isolating of high-quality, intact RNA from strawberry leaves of Xanthomonas fragariae, three most suitable reference genes, crucial for the normalization of RT-qPCR data for this pathogen and accurate expression analysis of target genes. In our study, various mathematic algorithms: NormFinder geNorm, BestKeeper, the delta CT method, RefFinder were adopted for validation of most stable reference genes from nine candidate genes (ffh, glyA, gyrA, gyrB, proC, pykA, recA, rpoB, rpoD). The analyses allowing to select three most suitable pioneer reference genes, gyrB, ffh, and pykA, that we recommend for the normalization of RT-qPCR data and for the study of the expression of target genes in Xf. Moreover, their combination as references allowed for an accurate expression analysis and computation of the fold change of the flhF and iroN2 genes in Xf. These two genes are important for the success of the colonization of plant tissue and pathogenicity and sequences of primers designed to study these genes, are presented.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Xanthomonas fragariae (Xf) is a Gram-negative, slow-growing bacterium and a causal agent of angular leaf spots (ALS) on strawberries (Fragaria spp) worldwide [29]. In 1986, Xf was classified as a quarantine organism on the A2 list by the European and Mediterranean Plant Protection Organization (EPPO) as well as the European Union (EU) [9]. Currently, its phytosanitary categorization is EPPO A2 list no. 135, EU Annex II/A2. The natural hosts of Xf are Fragaria × ananassa, its parents Fragaria chiloensis and Fragaria virginiana, and various wild strawberry species [9]. Early disease symptoms of ALS are tiny, angular water-soaked areas on leaves, leading to the development of angular spots on the lower leaf surface, that enlarge over time, become reddish necrotic irregular spots on the upper leaf surface and become surrounded by a chlorotic halo. The spots are translucent when using transmitted light but are seen as dark green when using reflected light. Under favourable conditions—moist air and at a temperature of 20 °C, a yellow bacterial exudate is visible on the lower leaf surface and calyces [1, 23].
Occasionally, ALS may cause significant damage to stock of planting material as well as in strawberry production [35], leading to fruit yield losses of about 10% [51], however, usually crop losses are low. The occurrence of ALS was first reported in Minnesota, U.S.A. in 1959 [29]. Since 1970, ALS has been reported in many European countries and has spread globally [9, 30]. Reports on the newest occurrence of ALS are from China and Iran [27, 67]. A comparative genomics approach using genome sequences of different Xf strains has been published and indicate high homogeneity of the sequenced Xf strains, although some differences have been detected in plasmids [14, 18, 64]. The role of plasmids in the pathogenicity of Xf has not been documented.
Although colonization of and movement in strawberry of Xf have been described [66] and critical pathogenesis-related gene clusters have been reported for the Xanthomonas genus, e.g., those encoding the structural elements of the Type III Secretion System (T3SS) as well as the Type II Secretion System (T2SS) in Xf [14, 18, 64], nothing is known about the expression of bacterial genes during strawberry infection. Understanding the pathogenic process, including changes at the transcription level, is important from the cognitive point of view and it may lead to the development of new disease control strategies.
To study the expression differences of pathogen genes, molecular techniques, such as RNA sequencing (RNA-seq) based on next-generation sequencing (NGS) platforms or quantitative reverse transcription PCR (RT-qPCR), which require a good quantity and quality, of unimpaired RNA, are necessary. To analyze the bacterial transcriptome in plants, crucial steps have to be carried out: (i) correct inoculation of a host plant species and appropriate sampling methods; (ii) selection of adequate RNA isolation and purification methods for the studied organism, allowing for isolation of RNA that is intact and free of contamination. Although laboratory techniques for RNA isolation/extraction using a manual method have already been published and used [33, 37, 42, 52], commercial kits that allow for fast extraction of RNA to be used in the high-throughput analysis are most often used by scientists [22, 25]. From previously published studies, it is known that depending on the type of sample, different RNA extraction kits vary in terms of the yield, quality, and integrity of the obtained RNA [8, 21, 53]. Therefore, to avoid the influence of low-quality RNA on the results of downstream applications, scientists must perform additional work and consider adapting the methodology for the studied organism [19]. Independent of the RNA isolation method selected, it is necessary to consider the need to eliminate contaminating genomic DNA with DNase I, which can adversely affect the results of several applications. Because the majority of bacterial RNA in the bacterial cell is not polyadenylated, as it is in eukaryotes, an additional intermediate step to remove abundant rRNA transcripts from total RNA, which constitutes approximately 90% of the RNA species, is necessary for NGS/transcriptome analysis [6, 15, 55].
Currently, quantitative, reverse transcription PCR (RT-qPCR) is one of the most efficient and robust methods for studying gene expression differences between studied samples e.g. bacteria, plants and for validating data obtained from RNA-seq [5, 32, 42, 45, 49]. Although RT-qPCR is highly specific, sensitive, convenient and rapid in execution [3], it also has certain limitations and requires technical skill to attain precision in the pre-analytical steps, which can influence the Ct values, i.e., obtaining high quality RNA good reverse-transcription and PCR amplification as well as avoiding pipetting errors [21, 61, 63, 65]. Another issue is the minimization of the variation between samples. To achieve this, an appropriate choice of reference genes that are stably expressed under different environmental conditions for the normalization process is necessary [3, 16, 42]. However, up till now a universal set of genes for normalization doesn’t exist. Although a few reference genes, including those for 18S rRNA, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β-actin and tubulin, are often used to normalize gene expression for eukaryotic species, it has been reported in numerous studies that the expression levels of these housekeeping genes vary under changing conditions [54, 57, 59, 71]. Moreover, in prokaryotic species, there is no agreement regarding which genes can be used as a universal reference set. Additionally, there are no studies on the selection of the most suitable reference genes in Xf. Studying the expression of this pathogen genes would be very interesting in the context of better understanding the pathogenic mechanism. For validation, several mathematic algorithms, which are commonly applied for data normalization [2, 44, 58, 63, 70], have been developed and are supported by the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) Guidelines for more accurate and error-free Analysis of Expression [4, 5].
The purposes of our study were: (i) to select the most efficient method of inoculating strawberry with Xf; (ii) to select the optimal RNA extraction method in terms of its ability to obtain high quantity, pure total RNA and intact mRNA from Xf in planta; and (iii) to determine the suitability and validate the most appropriate Xf reference genes to determine quantitative level of expression of the bacterial genes flhF and iroN2 which are important for the successful colonization of plant tissue and pathogenicity after its inoculation of strawberry leaves employing different mathematic algorithms.
Materials and methods
Inoculation methods
Young potted plants of strawberry cv. Elsanta, after the full development of three true leaves, were inoculated under greenhouse conditions with the Xanthomonas fragariae (Xf) strain IPO 3485 (IPO = Instituut voor Plantenziektenkundig Onderzoek) isolated in 2011 in the Netherlands, in late spring (May/June). To check the possibilities of successful plant inoculation and symptoms development different methodologies were used. Initially, in the first trial, which was conducted for 4 weeks, the conditions were: 20–25 °C with a relative humidity (RH) of 30–50% under natural light and naturally maintaining humidity conditions. For inoculations of strawberry plants, we used four different methods, each in two versions, the latter being (a) covering the plants with plastic bags for 48 h to maintain the high-humidity conditions in the first phase of infection (2 days) and (b) not covering the plants and maintaining the above mentioned greenhouse conditions. The four different methods were: (1) high-pressure spraying of the abaxial and adaxial surfaces of leaves with a 106 cfu mL−1 bacterial suspension in sterile water of a 48 h culture, grown on Wilbrink’s medium [31]; (2) leaf infiltration with a 106 cfu mL−1 bacterial suspension using a syringe; (3) treatment of the strawberry leaf surface with carborundum powder and low-pressure spraying of the abaxial and adaxial surfaces of leaves with a 106 cfu mL−1 bacterial suspension in sterile water of a 48 h culture, grown on Wilbrink’s medium; and (4) treatment of the strawberry leaf surface with carborundum and high-pressure spraying on both sides of leaves with a 106 cfu mL−1 bacterial suspension. In the second trial, we tested all inoculation methods described above, but in only one version—plants were not covered with plastic bag—to increase low RH in the greenhouse we ‘built ‘special plastic tent which allows keeping higher humidity level throughout the duration of the experiment. In the second trial, plants were maintained in a quarantine greenhouse at higher temperature 25–30 °C with an RH level of 70–90% under natural light conditions and were systematically watered. Negative control plants were treated similarly, however instead of the bacterial suspension, sterile distilled water was used. RNA isolation was performed to choose the best method for plant inoculation based on the RNA amount obtained. Each technique was tested on 2 sets of 3 plants each. The development of the disease was observed within 4 weeks after inoculation.
Total Isolation of total RNA, DNase treatment and depletion of rRNA for NGS
For total RNA extraction from the Xf pure culture strain IPO 3485 grown overnight in liquid Wilbrink’s medium [31] and from diseased plant material prepared as above, two commercial kits, were used: (1) the TRIzol® Max™ Bacterial RNA Isolation Kit with own modifications, and (2) the Total RNA Purification Kit (Norgen Biotek, Thorold, ON, Canada) according to the manufacturer’s instructions.
For inoculated plant material 10, 15 and 18 days after inoculation (25–30 °C, RH 70–90%), samples were processed and total RNA was isolated. At each time point, RNA was isolated separately from the leaves of each sample inoculated by the four different methods used.
In our first trials, RNA isolation from bacteria in planta, independent of the inoculation method (size/amount of symptoms) and RNA kit used, the amount of bacterial RNA obtained was very low (only a few to approx. 15 ng/µL), although a few independent repetitions of isolation were performed. Therefore, the RNA isolation step had to be preceded by additional procedures to better release bacteria from the plant material. Leaves with disease symptoms were collected, weighed (from 1.5 to 2.5 g were taken), cut into fragments and immediately incubated with RNAlater® Stabilization Solution (ThermoFisher Scientific) for 25 min with shaking in a Excella E24 Incubator Shaker Series (New Brunswick Scientific, Edison, New Jersey, USA) at 26 °C. Subsequently, the mixture was centrifuged, and the pellet was suspended in nuclease-free, sterile H2O. The amount of water added depended on the weight of the plant material used for RNA isolation and was calculated according to: 2.5 ml of water per 0.5 g of leaf material. Then, the Genomic Mini AX SOIL Spin kit (A&A Biotechnology, Gdynia, Poland), which has high efficiency for isolating genomic DNA from soil, was used. Briefly, the water-leaf-bacteria mixture was vortexed for 30 s at RT and incubated on ice for 5 min. During the incubation time, 1500 μL or more (depending on the weight of the plant material, 500 μL per 0.5 g of material) of a so-called R separation solution (Genomic Mini AX SOIL Spin, A&A Biotechnology, Gdynia, Poland) was poured into a 15 ml Falcon tube and kept on ice for 5 min. The water-leaf-bacteria mixture was slowly and carefully poured over the R solution layer and the tube was carefully placed in a tilting rotor and centrifuged at 4500×g for 10 min. The entire supernatant (two layers) was transferred to a new 15 ml Falcon tube, and nuclease-free H2O was added up to 30 ml or more (depending on the weight of the plant material and buffer amounts used in the procedure, up to 10 ml per each 0.5 g of material); then, the mixed sample was centrifuged at 4500×g for 10 min. The supernatant was poured out carefully, and the pellet was used for the total RNA isolation procedure with the selected kit.
For the removal of DNA contamination, what is essential for downstream applications, two commercial DNAse were used: DNase I (ThermoScientific) and the TURBO DNA-free™ Kit (Life Technologies). The efficiency of removing DNA was tested by PCR with the primers 245A and 245B [46], which are complementary to the genome of Xf, preceded by the use of a reverse transcription kit, the iScript cDNA synthesis kit (Biorad, Hercules, CA). The obtained RNA free from DNA was assessed according to the RNA integrity number (RIN) [56] where 10 is recognized to be the most unimpaired pure RNA and 1-the lowest value means the most degraded RNA sample [38]. Determination of the RIN, quality and concentration was performed on an Agilent 2100 Bioanalyser (Agilent Technologies, Palo Alto, CA) using the Agilent RNA 6000 Nano LabChip® kit. The procedure according to the manufacturer’s instructions was used. Based on our previous experience focused on obtaining of high quantity and quality of bacterial mRNA for next-generation sequencing, depletion of rRNA is an essential step. Therefore, based on our previous experience with removing of rRNA from Xf RNA samples, the Ribo-Zero™ Magnetic Kit (Gram-Negative Bacteria, http://www.illumina.com) was used according to the manufacturer’s instructions.
Candidate reference gene selection and primer design
Based on a screen of literature data concerning the most often used genes for validation as reference ones in other bacterial species [7, 12, 26, 36, 50], nine reference genes i.e. gyrB (DNA gyrase B), ffh (signal recognition particle protein), glyA (serine hydroxymethyltransferase), gyrA (DNA gyrase A), pykA (pyruvate kinase), proC (pyrroline-5-carboxylate reductase), recA (recombinase A), rpoB (DNA-directed RNA polymerase subunit beta) and rpoD (encoding sigma factor 70), were used for determination of their usefulness as reference for the study of virulence gene expression in Xf. To minimize the possibility of co-regulation between the genes, candidates with various biological functions were chosen. Primers were designed with the PrimerSelect program of the LASERGENE package (DNASTAR) based on the gene sequences of strain Xf IPO 3485 (= NBC 2815; NBC = Naktuinbouw Bacterial Collection) genome (LT853880, LT853881) (Gétaz et al. 2017). The selected primers were synthesised by Genomed S.A. (Warszawa, Poland). The specificity of the designed primers and their usefulness were examined twice using conventional and real-time PCR in the manner described by Kałużna et al. [26].
Reverse transcription, qPCR and determination of PCR efficiency
From the total RNA, 150 ng/µL was used for cDNA synthesis by applying an iScript cDNA synthesis kit (Biorad, Hercules, CA). The reaction was conducted in a thermocycler Bio-Rad CFX96 using SsoAdvanced SYBR Green Supermix (Bio-Rad, Hercules, CA). The prepared reaction mixture (20 μL in total volume), contained: 15 ng of cDNA, 1 × SYBR Green Supermix and 0.5 mM of each primer pair (Table 1) performed in a separate PCR for each gene. Nuclease free water was used as negative control. The PCR conditions used were as previously described [26] with slight modification, including the use of temperature of 60 °C instead of 59 °C. The amplification efficiency of each reference gene was determined and calculated as described previously [26].
Expression data and stability of the reference genes
The data concerning expression for the reference genes were calculated as the quantification cycle (Cq) values obtained from the RNA originating from a pure culture and the RNA of bacteria in planta. Five programs as in our previous paper [26], NormFinder [2], geNorm [63], BestKeeper [44], the delta CT method [58] and the RefFinder [70] were used to determine the level of stability of the candidate reference genes. The methodology of data preparations and Cq conversions for particular programs and formula used for efficacy calculations are described in Kałużna et al., [26].
Expression analysis of the pathogenicity—related genes
For expression profile analysis, the following target genes, the products of which are important for the successful colonization of plant tissue and pathogenicity were selected: flhF, which is a flagellar biosynthesis regulator, and iroN2, which is the TonB-dependent receptor. Although the latter does not play a direct role in pathogenesis it allows Gram-negative bacteria to take up scarce resources from nutrient-limiting environments as well as siderophores, heme etc., [17, 40]. The primers for the target genes (Table 1) were designed using the PrimerSelect program of the LASERGENE package (DNASTAR) and synthesized by Genomed S.A. (Warszawa).
In order to compute the relative fold changes in gene expression of the studied genes,data from Cfx96 (Bio-Rad, Hercules, CA) were analysed using the comparative 2−ΔΔCt method [34] was used.
Results
Strawberry plant inoculation
Comparing the inoculation efficiency under two temperature/humidity conditions, earlier and more intensive symptoms were observed at higher temperatures 25–30 °C and higher humidity 70–90%. In the temperature range 20–25 °C and at the RH 30–50%, the first symptoms, necrotic lesions were observed quite late, at 14 days after inoculation, on strawberry plants inoculated by infiltration with a syringe. In the case of higher temperature/humidity, the symptoms appeared earlier, with the first symptoms observed 7–10 days after inoculation of plants sprayed under high pressure and after infiltration. Similar symptoms were observed in the case of applying of low pressure but with less intensive symptom formation.
The lesions size slightly increased throughout the experiment with 1–2 mm and a few additional spots developed due to the secondary spread of the bacteria. In the infiltration method, experiments apart from a water soaked tissue, necrosis of leaf tissue at the infiltration location was also observed (Fig. S1). Disease symptoms were not observed in non-inoculated control plants.
Total RNA isolations, DNase treatment and depletion of rRNA
The quality and quantity of RNA obtained from individual samples largely depended on the method of inoculation. The analysis of the total RNA concentration and integrity (RIN parameter) showed that, the highest concentration (> 2 µg μL−1) and best quality RNA (RIN 9.8) were obtained from plants, 15 days after inoculation by spraying under pressure (kept at 25–30 °C with a humidity level from 70–90%) and using total RNA Purification Kit (Norgen Biotek) proceeded by the Genomic Mini AX SOIL Spin kit (A&A Biotechnology, Gdynia, Poland). For pure culture cells of the Xf strain IPO 3485 the RIN was 10. When low-pressure spraying was applied, lower concentrations and slightly lower RIN values (lower of 0.5–0.7 lower) were recorded. When using the TRIzol® Max™ Bacterial RNA Isolation Kit, the RIN was approximately 7–8. Smaller amounts and bad quality RNA were obtained from leaf samples infiltrated by a syringe (RIN < 5). This could possibly be due to the dark brown necrotic spots formation in the infiltration points where a rapid host cell death likely leads to activity of reactive oxygen species (ROS) causing some degree of RNA degradation.
Both DNases used were equally effective for the removal of genomic DNA; however, we chose DNase I for our study (ThermoScientific). The Ribo-Zero™ Magnetic Kit used to obtain mRNA for next-generation sequencing allowed us to obtain a good quantity of 30–70 ng/µL of Xf mRNA from all samples.
Selection of reference genes from candidates and design of the primer
All nine candidate reference genes were used to further examination of gene expression stability and their ranking based on the data from conventional and real-time PCRs. All the data concerning the name of genes used, forward and reverse primer sequences, length of amplicons, melting temperatures (Tm), PCR efficiency, and regression coefficients obtained are listed in Table 1. Single product with the expected size from 72 to 150 bp was obtained with the primer pairs designed for each gene and used in PCRs for all genes (Table 1, Fig. S2). The verification of primers by real-time PCR gave melting curves with a single peak, where the Tm varied from 86 to 90.50 °C and no unexpected or additional peaks were noticed (Fig. S3). This confirmed that the primers were not able to form dimers. The determined efficiencies of the primers designed for all candidate reference genes ranged from 93.1 to 109.2% for all genes depend on each primer pairs/genes. The linear correlation coefficients (r2) were from 0.989 to 0.999 (Table 1).
Stability and expression levels of the candidate reference genes
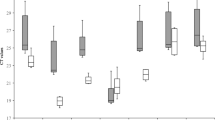
Cq values obtained (Fig. 1) for the candidate reference genes were quite variable, indicating varied expression. Therefore, statistical tools were necessary to estimate the stability of genes studied allowing us to determine the most appropriate references for studies of gene expression (Fig. 1). The Cq values obtained from all candidates varied from 20.17 to 31.25. The median and average Cq values varied for all candidate genes studied. The rpoD gene had the lowest mean value (24.24), while proC the highest one (29.57).
Stability of expression of the reference genes
Statistical algorithms applied in geNorm, NormFinder, BestKeeper, the delta-Ct method, and the RefFinder web-based tool programs, were used to determine the stability of the expression of the candidate reference genes (Table 2).
Based on the geNorm analysis, gyrB and ffh genes followed by gyrA and rpoB genes possess the highest stability as they showed the lowest M-values (0.697, 0.740, 0.749 and 0.834, respectively) (Table 2). RpoD turned out to be the least stably expressed gene. Based on the geNorm analysis, the gyrB and ffh followed by gyrA and rpoB genes had the lowest M-value (0.697, 0.740, 0.749 and 0.834, respectively), what pointing out that these genes have the highest stability (Table 2). Only rpoD showed an M-value above the determined default limit of M < 1.5 and therefore turned out to be the least stably expressed gene.
The analysis of V-value—pairwise variation determined for the reference genes studied showed that value V4/5 was below the default threshold value of 0.15
The Normfinder analysis indicated that the most stably expressed genes were gyrB, ffh, rpoB and gyrA (SV: 0.208, 0.244, 0.268 and 0.299, respectively). rpoD and recA were the least stably expressed genes findings similar to those obtained by geNorm (Table 2). Additionally, it was shown that gyrA and gyrB are the best combinations of genes for further studies of the expression of interest genes (calculated stability value 0.132).
The results obtained by BestKeeper analysis was different from both: GeNorm and Normfinder and revealed that all genes can be treated as inconsistent as reference genes for RT-qPCR because they have an SD value calculated greater than 1. Of all analyzed candidates the genes rpoD and gyrA were designated as the most stably expressed with SD: 1.08 and 1.74, respectively followed by pykA gyrB and ffh (SD: 1.85, 2.01 and 2.07, respectively) (Table 2). glyA was found to be the least stably expressed gene.
According to the delta Ct method genes gyrB, pykA and ffh, (SD: 0.46, 0.52, 0.54) were the most stably expressed ones; and rpoD the least stably expressed gene, with an SD of 1.27 (Table 2).
Based on the RefFinder web-based tool, the ranking of the candidate reference genes was similar to those obtained by geNorm and NormFinder. The results of BestKeeper were similar when using RefFinder, although the efficiency value was not included as in the original BestKeeper software. The complete ranking from the RefFinder was as follows: the gyrB, ffh, pykA and gyrA (GM: 1.41, 2.78, 2.78, 3.81, respectively) were the most stably expressed genes, and recA and proC were the least stably expressed (Table 2; Table S1).
Expression analysis of the flhF and iroN2
Based on the analysis of the gene expression differences of the flhF and iroN2 genes normalized with gyrB, ffh, and pykA as the selected reference genes, flhF was up-regulated in planta; however, iroN2 was down-regulated. The flhF gene was up-regulated by 231.64-fold (SE 42.01) compared with that of a pure bacterial culture; however, for the iroN2 gene, the opposite result was noted i.e., down-regulation in planta (− 16.2-fold; SE 2.072) compared to pure bacterial culture. Compared to the results of transcriptome analysis by RNA seq, the expression values of the flhF and iroN2 genes obtained in this study were similar [48].
Discussion
Xanthomonas fragariae causes damages to plant stock as well as strawberry production worldwide. Therefore, identification of its pathogenicity factors would be of great value from cognitive and practical points of view. Several pathogenesis-related gene clusters have already been described [14, 18, 64], and expression analysis of these clusters in different stages of the infection process would be valuable. The present study is one of a few works focused on the selection and validation of bacterial reference genes for the normalization of RT-qPCR gene expression [20, 26, 50, 62, 69]. To the best of our knowledge, no specific internal references for Xf have been described. Normalization of the expression level of the studied genes, against validated, stably expressed reference genes allow to avoid the obtaining of false positive/false-negative results. It also can compensate and equalize variations that can arise due to factors e.g. RNA quality of samples analyzed, cDNA synthesis. Therefore, this study constitutes the first work on the validation of candidate reference genes using a numerous algorithms for use for the relative quantification of flhF and iroN—target gene expression in Xf. RT-qPCR has recently become the method of choice for analyses, allowing the determination of the different expression changes of target genes relative to reference genes [45], because it is simple, reproducible and sensitive [3]. However, RT-qPCR requires technical skills e.g. precise in pipetting) and the development of a few analytical steps before expression study e.g. crucial normalization and validation assays.
Our methodology allowed the selection of the most suitable inoculation method of strawberry leaves with Xf based on analyses of four methods of obtaining high-quality RNA, which appeared to be strictly depended on the method of inoculation.
The first disease symptoms appeared in plants inoculated with infiltration 25–30 °C; 70–90% RH. In the case of the pathogenicity tests performed by Kastelein et al. [28], the selected optimal temperature differed from our—it was lower and ranged from 17–20 °C and 80% RH, it gave them under this condition and 10% of the diseased leaves already diseased 7 days post inoculation. Early symptoms of infection in angular water-soaked lesions on the abaxial surface of leaves appeared approximately 8 days after inoculation in the study of Allan-Wojtas et al. [1], they also used lower temperatures 18–22 °C and high relative humidity (> 95%). Wang et al. (1966) inoculated plants with Xf100-GFP mutant strain at 25 °C and 60% relative humidity during the day and 15 °C and 90% RH during the night [66 and first lesions as in our work appeared 7 days post-inoculation.In our case, covering plants for 48 h with plastic bags after inoculation to maintain high humidity but under lower temperature conditions did not positively influence the speed of disease symptom development, and moreover, covering with plastic bags caused significant infestation by fungal pathogens, which interfered with bacterial RNA isolation (data not shown). In Kastelein et al. [28] and Allan-Wojtas et al. [1], air-pressurized spraying onto the abaxial surface of leaves was applied as a successful method of inoculation, similar to the methodology used in our paper. The methodology applied (°C and RH) and results obtained—symptoms appearing—in presented above studies suggest that besides the obvious essential high humidity conditions the use of an optimum temperature is also crucial for the bacteria multiplication and early symptoms development. However, in the work of Kastelein et al. [28] 20 °C was used throughout the entire experiment, and the results anyway appeared quite early. Additionally, similar inoculation methods, which were found in our case to be best for inoculation, such as high-pressure spraying on both sides of leaves with a bacterial suspension in sterile water, were used for pathogenicity tests that were performed to confirm the pathogenicity of the pathotype strain of Xanthomonas arboricola pv. fragariae (another pathogen of strawberry [10]). In they work only slight differences in symptoms development were observed for inoculations using low-pressure spraying of the bacterial suspension (107 cfu mL−1) on the lower surface of leaves [10].
The highest concentration and best quality of total RNA were obtained from plant samples inoculated by spraying under pressure, 15 days after inoculation that were kept at 25–30 °C with a humidity level 70–90%. During the first attempts, the main obstacle was the inability to isolate sufficient quantities of bacterial RNA. In the first few trials we used a procedure that worked well for another bacterium, Erwinia amylovora in apple shoots [25], but in the case of Xf, the efficacy of RNA isolation was much lower. We supposed that this failure was caused by the inefficient release of bacterial cells from plant tissue. To overcome this problem, the most crucial step of the total RNA isolation procedure of Xf from strawberry leaves consisted of a combination of different systems, namely, the partial adoption of the reagents and methodology of the Genomic Mini AX SOIL Spin (A&A Biotechnology) kit followed by application of the total RNA Purification Kit (Norgen Biotek). Our study demonstrated once more that there is no common isolation method for all types of microorganisms (especially bacteria for different plant material or when used in different environments; thus there is a need to develop the protocol for an individual need and purpose [41, 42]. We assume that the procedure we developed can work for other bacteria, particularly in cases in which the extraction of bacterial cells from a particular tissue is difficult, but that also has to be tested in each individual host–pathogen combination.
From the literature, it is known and recommended that the minimum RNA integrity number (RIN) for analyses should be at least seven; higher than eight is considered to be optimal [11, 24]. Here, we obtained RNA with a RIN near or equal to 10 in all samples. However, so high RINs were not obtained in other bacterial species, such as Erwinia amylovora, because apparently the ribosomal RNA ratio is disturbed due to a 99-bp intervening sequence (IVS) insertion within the 23S rRNA gene [47, 60]. It is known from the literature that IVSs are transcribed but next finally removed by RNase III, resulting in fragmented rRNA [47]. The presence of two extra peaks indicates the presence of these fragmented rRNAs, the latter making it impossible to obtain the maximum RIN value.
In our study, nine reference housekeeping genes of Xf were chosen and their expression determined. The amplification efficiency of the designed primers for these genes was calculated using the conventional standard curve method. We preferred this method, although it has some disadvantages (it is time-consuming, and requires production of repeatable and reliable standards [43, 72]; its efficiency of reaction differs from the relatively stable in the exponential phase to gradually decreasing later [68]; the efficiency calculated can be over-estimated [50]). This method was also found to be reliable and preferred in our previous expression study and other recently published papers [26, 39, 42, 54].
To evaluate the candidate reference genes stability to use the most stably expressed for normalization of RT-qPCR data and for calculation of the expression of genes involved in pathogenicity and in playing a role in plant tissue colonization (flhF and iroN2), several different mathematical algorithms that have already been used by several authors were adopted [32, 39, 42, 50, 73]. Based on the results of the algorithms used, the comprehensive range demonstrated that gyrB, followed by ffh, pykA and gyrA, were the most stably expressed reference genes. According to the MIQE Guidelines, at least two reference genes are acceptable and required to determine gene expression changes; however, three reference genes are preferred, and as stated by authors the use of only one is not acceptable [5]. The results of the expression analysis using three reference genes allow to obtaining more precise and trustworthy results. It has been reported that the use of a single reference gene can generate up to a 20-fold error in the expression data [59, 63].
The gyrB gene was also identified as the most stably expressed gene in Aeromonas salmonicida subsp. salmonicida [50] and Staphylococcus pseudintermedius [7]; however, it was expressed at varying levels under different experimental conditions in the validation of the reference genes for RT-qPCR analysis in Burkholderia pyrrocinia [69]. GyrA and ffh were the most stable reference genes in normalization studies of reference genes in Azospirillum brasilense [36] and Pectobacterium atrosepticum [62], respectively. RpoB was present in a group of the most stably expressed genes [20] among candidates selected in studies on Xanthomonas citri subsp. citri genes during the infection of Citrus sinensis. In our study, rpoB was also one of the most stably expressed genes. The rpoD gene was identified as one of three genes whose expressions were stable throughout the growth of Gluconacetobacter diazotrophicus strain PAL5 on different carbon sources [13], while in our case, and also for Aeromonas salmonicida subsp. salmonicida [50], this gene was the least stably expressed gene. The variation in the results obtained in many papers for a particular organism and environment applied, point at the necessity to perform gene normalization for each environment and species.
When analysing flhF and iroN2, it was found that flhF, a flagellar biosynthesis pathway gene, was highly up-regulated, while iroN2, which encodes the TonB-dependent receptor, was down-regulated in planta. The results of the reference gyrB, ffh and pykA genes in terms of the relative gene expression analysis agree with the results obtained in preliminary analysis of the expression of flhF and iroN2 in transcriptomes obtained after RNA-seq [48]. GyrA did not make significant changes in the results obtained (data not shown). Moreover, as was the case in the study of Rivera et al. [50], geNorm analysis showed that at least five reference genes should be used in all studies. In our hands, however, the use of three, for more genes, did not make meaningful changes in the results.
Conclusions
We present in this article the research that led to the development of a novel, procedure for the isolation of high-quality, intact RNA of Xanthomonas fragariae in planta. The described unique methodology can be adopted for other bacteria, in each individual host–pathogen combination. Also, our results provide the designed and validated primers for the group of reference genes which can be useful for other studies of gene expression analysis of Xanthomonas fragariae. Numerous programs and algorithm analyses enabled the selection and first-time use of three the most suitable reference genes, gyrB, ffh, and pykA, that are crucial for the normalization of RT-qPCR data. The adopted genes used for the normalization of RT-qPCR data allowed an accurate expression analysis and computation of the fold change of the flhF and iroN2 genes which play a role in the successful colonization of plant tissue and pathogenicity by Xf.
References
Allan-Wojtas P, Hildebrand PD, Braun PG, Smith-King HL, Carbyn S, Renderos WE (2010) Low temperature and anhydrous electron microscopy techniques to observe the infection process of the bacterial pathogen Xanthomonas fragariae on strawberry leaves. J Microsc 239:249–258. https://doi.org/10.1111/j.1365-2818.2010.03373.x
Andersen CL, Jensen JL, Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a modelbased variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250
Bustin SA (2002) Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol 29(1):23–39
Bustin SA, Beaulieu JF, Huggett J, Jaggi R, Kibenge FS, Olsvik PA, Penning LC, Toegel S (2010) MIQE precis: practical implementation of minimum standard guidelines for fluorescence-based quantitative real-time PCR experiments. BMC Mol Biol 11(1):74
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT (2009) The MIQE Guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55(4):611–622
Chen Z, Duan X (2011) Ribosomal RNA depletion for massively parallel bacterial RNA-sequencing applications. Methods Mol Biol 733:93–103
Crawford EC, Singh A, Metcalf D, Gibson TW, Weese SJ (2014) Identification of appropriate reference genes for qPCR studies in Staphylococcus pseudintermedius and preliminary assessment of icaA gene expression in biofilm-embedded bacteria. BMC Res Notes 7:451
Deng MY, Wang H, Ward GB, Beckham TR, McKenna TS (2005) Comparison of six RNA extraction methods for the detection of classical swine fever virus by real-time and conventional reverse transcriptase-PCR. J Vet Diagn Invest 17:574–578
EPPO (2006) Xanthomonas fragariae. EPPO Bulletin 36:135–144
Ferrante P, Scortichini M (2018) Xanthomonas arboricola pv. fragariae: a confirmation of the pathogenicity of the pathotype strain. Eur J Plant Pathol 150(3):825–829
Fleige S, Pfaffl MW (2006) RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med 27:126–139
Florindo C, Ferreira R, Borges V, Spellerberg B, Gomes JP, Borrego MJ (2012) Selection of reference genes for real-time expression studies in Streptococcus agalactiae. J Microbiol Methods 90(3):220–227
Galisa SP, da Silva HA, Macedo AV, Reis VM, Vidal MS, Baldani JI, Simões-Araújo JL (2012) Identification and validation of reference genes to study the gene expression in Gluconacetobacter diazotrophicus grown in different carbon sources using RT-qPCR. J Microbiol Methods 91(1):1–7
Gétaz M, van der Wolf JM, Blom J, Pothier JF (2017) Complete genome sequences of three isolates of Xanthomonas fragariae, the bacterium responsible for angular leaf spots on strawberry plants. Genome Announc 5(32):e00642
Giannoukos G, Ciulla DM, Huang K, Haas BJ, Izard J, Levin JZ, Livny J, Earl AM, Gevers D, Ward DV, Nusbaum C, Birren BW, Gnirke A (2012) Efficient and robust RNA-seq process for cultured bacteria and complex community transcriptomes. Genome Biol 13(3):r23
Gutierrez L, Mauriat M, Guénin S, Pelloux J, Lefebvre J, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C, Van Wuytswinkel O (2008) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction RT-PCR analysis in plants. Plant Biotechnol J 6:609–618
Harshey RM (2003) Bacterial motility on a surface: many ways to a common goal. AnnuRev Microbiol 57:249–273
Henry PM, Leveau JHJ (2016) Finished genome sequences of Xanthomonas fragariae, the cause of bacterial angular leaf spot of strawberry. Genome Announc 4(6):e01271
Imbeaud S, Graudens E, Boulanger V, Barlet X, Zaborski P, Eveno E, Mueller O, Schroeder A, Auffray C (2005) Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Res 33(6):e56
Jacob TR, Laia ML, Ferro JA, Ferro MI (2011) Selection and validation of reference genes for gene expression studies by reverse transcription quantitative PCR in Xanthomonas citri subsp citri during infection of Citrus sinensis. Biotech Lett 33(6):1177–1184
Jahn CE, Charkowski AO, Willis DK (2008) Evaluation of isolation methods and RNA integrity for bacterial RNA quantitation. J Microbiol Methods 75:318–324
Jain N, Vergish S, Khurana JP (2018) Validation of house-keeping genes for normalization of gene expression data during diurnal/circadian studies in rice by RT-qPCR. Sci Rep 8(1):3203
Janse JD, Rossi MP, Gorkink RFJ, Derks J, Swings J, Janssens D, Scortichini M (2001) Bacterial leaf blight of strawberry (Fragaria ananassa) caused by a pathovar of Xanthomonas arboricola, not similar to Xanthomonas fragariae Kennedy & King. Description of the causal organism as Xanthomonas arboricola pv. fragariae (pv. nov., comb. nov.). Plant Pathol 50:653–665
Jeffries MKS, Kiss AJ, Smith AW, Oris JT (2014) A comparison of commercially-available automated and manual extraction kits for isolation of total RNA from small tissue samples. BMC Biotechnol 14:94
Kałużna M, Kuras A, Mikiciński A, Pulawska J (2016) Evaluation of different RNA extraction methods of high quality total RNA and mRNA from Erwinia amylovora in planta. Eur J Plant Pathol 146:893–899
Kałużna M, Kuras A, Puławska J (2017) Validation of reference genes for the normalization of the RT-qPCR gene expression of virulence genes of Erwinia amylovora in apple shoots. Sci Rep 7(1):2034
Kamangar SB, Van Vaerenbergh J, Kamangar S, Maes M (2017) First report of angular leaf spot on strawberry caused by Xanthomonas fragariae in Iran. Plant Dis 101(6):1031
Kastelein P, Krijger M, Czajkowski R, van der Zouwen PS, van der Schoor R, Janlik H, van der Wolf JM (2014) Development of Xanthomonas fragariae populations and disease progression in strawberry plants after spray-inoculation of leaves. Plant Pathol 63(2):255–263
Kennedy B, King T (1962) Angular leaf spot of strawberry caused by Xanthomonas fragariae sp. nov. Phytopathology 52:873–875
Kim DR, Gang GH, Jeon CW, Kang NJ, Lee SW, Kwak YS (2016) Epidemiology and control of strawberry bacterial angular leaf spot disease caused by Xanthomonas fragariae. Plant Pathol J 32(4):290
Koike H (1965) The aluminum-cap method for testing sugarcane varieties against leaf scald disease. Phytopathology 55:317–319
Koramutla MK, Aminedi R, Bhattacharya R (2016) Comprehensive evaluation of candidate reference genes for qRT-PCR studies of gene expression in mustard aphid, Lipaphis erysimi (Kalt). Sci Rep 6:25883
Leite GM, Magan N, Medina Á (2012) Comparison of different bead-beating RNA extraction strategies: an optimized method for filamentous fungi. J Microbiol Methods 88(3):413–418
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25:402–408
Maas JL (1998) Angular leaf spot. In: Maas JL (ed) Compendium of strawberry diseases, 2nd edn. American Phytopathological Society, St. Paul, pp 16–17
McMillan M, Pereg L (2014) Evaluation of reference genes for gene expression analysis using quantitative RT-PCR in Azospirillum brasilense. PLoS ONE 9(5):e98162
Mehra M (1996) RNA isolation from cells and tissues. In: Krieg PA (ed) A Laboratory Guide to RNA: isolation, analysis, and synthesis. Wiley, New York, pp 1–20
Mueller O, Lightfoot S, Schroeder A (2004) RNA Integrity Number (RIN)—Standardization of RNA quality control. Agilent Technologies, Technical Rep 5989-1165EN
Nakamura AM, Chahad-Ehlers S, Lima ALA, Taniguti CH, Sobrinho I Jr, Torres FR, de Brito RA (2016) Reference genes for accessing differential expression among developmental stages and analysis of differential expression of OBP genes in Anastrepha obliqua. Sci Rep 6:17480
Noinaj N, Guillier M, Barnard TJ, Buchanan SK (2010) TonB-dependent transporters: regulation, structure, and function. Annu Rev Microbiol 64:43–60
Nolan T, Hands RE, Bustin SA (2006) Quantification of mRNA using real-time RTPCR. Nat Protoc 1:1559–1582
Petriccione M, Mastrobuoni F, Zampella L, Scortichini M (2015) Reference gene selection for normalization of RT-qPCR gene expression data from Actinidia deliciosa leaves infected with Pseudomonas syringae pv. actinidiae. Sci Rep 5:16961
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45
Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: bestKeeper—excel-based tool using pair-wise correlations. Biotech Lett 26:509–515
Pfall MW (2006) Relative quantification. In: Dorak MT (ed) Real-time PCR, vol 63. Taylor and Francis Group, Abingdon, p 82
Pooler MR, Ritchie DF, Hartung JS (1996) Genetic relationships among strains of Xanthomonas fragariae based on random amplified polymorphic DNA PCR, repetitive extragenic palindromic PCR, and enterobacterial repetitive intergenic consensus PCR data and generation of multiplexed PCR primers useful for the identification of this phytopathogen. Appl Environ Microbiol 62(9):3121–3127
Pronk LM, Sanderson KE (2001) Intervening sequences in rrl genes and fragmentation of 23S rRNA in genera of the family Enterobacteriaceae. J Bacteriol 183:5782–5787
Puławska J, Kałużna M, Warabieda W, Mikiciński A (2017) Comparative transcriptome analysis of a lowly virulent strain of Erwinia amylovora in shoots of two apple cultivars—susceptible and resistant to fire blight. BMC Genomics 18:868
Reddy DS, Bhatnagar-Mathur P, Reddy PS, Sri Cindhuri K, Sivaji Ganesh A, Sharma KK (2016) Identification and validation of reference genes and their impact on normalized gene expression studies across cultivated and wild Cicer Species. PLoS ONE 11(2):e0148451
Rivera L, López-Patiño MA, Milton DL, Nieto TP, Farto R (2015) Effective qPCR methodology to quantify the expression of virulence genes in Aeromonas salmonicida subsp. salmonicida. J Appl Microbiol 118(4):792–802
Roberts P, Berger R, Jones J, Chandler CK, Stall RE (1997) Disease progress, yield loss, and control of Xanthomonas fragariae on strawberry plants. Plant Dis 81:917–921
Rubio-PiñaJA Zapata-PérezO (2011) Isolation of total RNA from tissues rich in polyphenols and polysaccharides of mangrove plants. Electron J Biotechnol 14:5
Rump LV, Asamoah B, Gonzalez-Escalona N (2010) Comparison of commercial RNA extraction kits for preparation of DNA-free total RNA from Salmonella cells. BMC Res Notes 3(1):211
Sagri E, Koskinioti P, Gregoriou ME, Tsoumani KT, Bassiakos YC, Mathiopoulos KD (2017) Housekeeping in Tephritid insects: the best gene choice for expression analyses in the medfly and the olive fly. Sci Rep 7:45634
Sarkar N (1997) Polyadenylation of mRNA in prokaryotes. Annu Rev Biochem 66:173–197
Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T (2006) The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol 7:3
Shen GM, Jiang HB, Wang XN, Wang JJ (2010) Evaluation of endogenous references for gene expression profiling in different tissues of the oriental fruit fly Bactrocera dorsalis (Diptera: Tephritidae). BMC Mol Biol 11(1):76
Silver N, Best S, Jiang J, Thein SL (2006) Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol 7:33
Singh S, Gupta M, Pandher S, Kaur G, Rathore P, Palli SR (2018) Selection of housekeeping genes and demonstration of RNAi in cotton leafhopper, Amrasca biguttula biguttula (Ishida). PLoS ONE 13(1):e0191116
Smits THM, Rezzonico F, Kamber T, Blom J, Goesmann A, Frey JE, Duffy B (2010) Complete genome sequence of the fire blight pathogen Erwinia amylovora CFBP 1430 and comparison to other Erwinia spp. Mol Plant Microbe In 23:384–393
Ståhlberg A, Hakansson J, Xian X, Semb H, Kubista M (2004) Properties of the reverse transcription reaction in mRNA quantification. Clin Chem 50:509–515
Takle GW, Toth IK, Brurberg MB (2007) Evaluation of reference genes for real-time RT-PCR expression studies in the plant pathogen Pectobacterium atrosepticum. BMC Plant Biol 7:50
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:5245–5250
Vandroemme J, Cottyn B, Baeyen S, De Vos P, Maes M (2013) Draft genome sequence of Xanthomonas fragariae reveals reductive evolution and distinct virulence-related gene content. BMC Genomics 14(1):829
Vermeulen J, De Preter K, Lefever S, Nuytens J, De Vloed F, Derveaux S, Hellemans J, Speleman F, Vandesompele J (2011) Measurable impact of RNA quality on gene expression results from quantitative PCR. Nucleic Acids Res 39:e63–e63
Wang H, McTavish C, Turechek WW (2018) Colonization and movement of Xanthomonas fragariae in strawberry tissues. Phytopathol 108(6):681–690
Wang J, Wei HL, Chang RK, Liu HQ, Wang YH (2017) First report of strawberry bacterial angular leaf spot caused by Xanthomonas fragariae in Tianjin, China. Plant Dis 101(11):1949–1949
Wong ML, Medrano JF (2005) Real-time PCR for mRNA quantitation. Biotechniques 39:75–85
Wu BY, Ye JR, Huang L, He LM, Li DW (2017) Validation of reference genes for RT-qPCR analysis in Burkholderia pyrrocinia JK-SH007. J Microbiol Methods 132:95–98
Xie F, Xiao P, Chen D, Xu L, Zhang B (2012) MiRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol Biol 80(1):75–84
Yuan M, Lu Y, Zhu X, Wan H, Shakeel M, Zhan S, Jin BR, Li J (2014) Selection and evaluation of potential reference genes for gene expression analysis in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae) using reverse-transcription quantitative PCR. PLoS ONE 9(1):e86503
Zhao S, Fernald RD (2005) Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol 12:1045–1062
Zhao X, Zhang X, Guo X, Li S, Han L, Song Z, Wang Y, Li J, Li M (2016) Identification and validation of reference genes for qrt-pcr studies of gene expression in Dioscorea opposita. BioMed Res Int BioMed Res Int 1:1–13
Acknowledgements
The authors would like to thank Dr. Sławomir Dąbrowski (A&A Biotechnology, Poland) for helpful advises on nucleic acid extraction and Mrs. Halina Kijańska for excellent technical help.
Funding
This study has received funding from the European Union Seventh Programme (FP7/2007–2013) under the grant agreement no 613678 (DROPSA). The research work was co-financed from financial resources for science in 2014–2018 granted for the implementation of an international co-funded project 3195/7.PR/2014/2.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Kałużna, M., Kuras, A. & Puławska, J. mRNA extraction of Xanthomonas fragariae in strawberry and validation of reference genes for the RT-qPCR for study of bacterial gene expression. Mol Biol Rep 46, 5723–5733 (2019). https://doi.org/10.1007/s11033-019-05006-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-019-05006-z