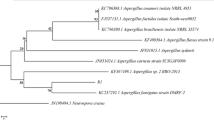
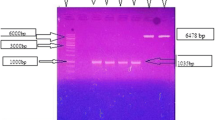
Abstract
Increased demand of enzymes for industrial use has led the scientists towards protein engineering techniques. In different protein engineering strategies, rational approach has emerged as the most efficient method utilizing bioinformatics tools to produce enzymes with desired reaction kinetics; physiochemical (temperature, pH, half life, etc) and biological (selectivity, specificity, etc.) characteristics. Xylanase is one of the widely used enzymes in paper and food industry to degrade xylan component present in plant pulp. In this study endo 1,4-β-xylanase (Xyl-11A) from Bacillus halodurans C-125 was cloned in pET-22b (+) vector and expressed in Escherichia coli BL21 (DE3) expression strain. The enzyme had Michaelis constant Km of 1.32 mg ml−1 birchwoodxylan (soluble form) and maximum reaction velocity (Vmax) 73.53 mmol min−1 mg−1 with an optimum temperature of 75 °C and pH 9.0. The thermostability analysis showed that enzyme retained more than 80% of its residual activity when incubated at 75 °C for 2 h. In addition, to increase Xyl-11A thermostability, an in-silico analysis was performedto identify the hot spot amino acid residues. Consensus-based amino acid substitution was applied to evaluate multiple sequence alignment of homologs and identified 20 amino acids positions by following Jensen-Shnnon Divergence method. 3D models of 20 selected mutants were analyzed for conformational transition in protein structures by using NMSim server. Two selected mutants T6K and I17M of Xyl-11A retained 40, 60% residual activity respectively, at 85 °C for 120 min as compared to wild type enzyme which retained 37% initial activity under same conditions, confirming the enhanced thermostability of mutants. The present study showed a good approach for the identification of promising amino acid residues responsible for enhancing the thermostability of enzymes of industrial importance.














Similar content being viewed by others
References
Busse-Wicher M, Li A, Silveira RL, Pereira CS, Tryfona T, Gomes TC, Skaf MS, Dupree P (2016) Evolution of xylan substitution patterns in gymnosperms and angiosperms: implications for xylan interaction with cellulose. Plant Physiol 171(4):2418–2431. https://doi.org/10.1104/pp.16.00539
Uday US, Choudhury P, Bandyopadhyay TK, Bhunia B (2016) Classification, mode of action and production strategy of xylanase and its application for biofuel production from water hyacinth. Int J Biol Macromol 82:1041–1054. https://doi.org/10.1016/j.ijbiomac.2015.10.086
Moreira LRS, Filho EXF (2016) Insights into the mechanism of enzymatic hydrolysis of xylan. Appl Microbiol Biotechnol 100(12):5205–5214. https://doi.org/10.1007/s00253-016-7555-z
Mechelke M, Koeck D, Broeker J, Roessler B, Krabichler F, Schwarz W, Zverlov V, Liebl W (2017) Characterization of the arabinoxylan-degrading machinery of the thermophilic bacterium Herbinix hemicellulosilytica—six new xylanases, three arabinofuranosidases and one xylosidase. J Biotechnol 257:122–130
Sermsathanaswadi J, Baramee S, Tachaapaikoon C, Pason P, Ratanakhanokchai K, Kosugi A (2017) The family 22 carbohydrate-binding module of bifunctional xylanase/β-glucanase Xyn10E from paenibacillus curdlanolyticus B-6 has an important role in lignocellulose degradation. Enzyme Microb Technol 96:75–84
Ribeiro AJM, Holliday GL, Furnham N, Tyzack JD, Ferris K, Thornton JM (2017) Mechanism and catalytic site atlas (M-CSA): a database of enzyme reaction mechanisms and active sites. Nucleic Acids Res 46(D1):D618–D623
Ang TF, Maiangwa J, Salleh AB, Normi YM, Leow TC (2018) Dehalogenases: from improved performance to potential microbial dehalogenation applications. Molecules. https://doi.org/10.3390/molecules23051100
Selvarajan E, Veena R (2017) Recent advances and future perspectives of thermostable xylanase. Biomed Pharmacol J 10(1):261–279
Jemli S, Ayadi-Zouari D, Hlima HB, Bejar S (2016) Biocatalysts: application and engineering for industrial purposes. Crit Rev Biotechnol 36(2):246–258. https://doi.org/10.3109/07388551.2014.950550
Shivange AV, Schwaneberg U (2017) Recent advances in directed phytase evolution and rational phytase engineering. In: Directed enzyme evolution: advances and applications. Springer, Cham, pp 145–172
Brown AW, Allison DB (2014) Using Crowdsourcing to evaluate published scientific literature: methods and example. PLoS ONE 9(7):e100647. https://doi.org/10.1371/journal.pone.0100647
Takami H, Nakasone K, Takaki Y, Maeno G, Sasaki R, Masui N, Fuji F, Hirama C, Nakamura Y, Ogasawara N, Kuhara S, Horikoshi K (2000) Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res 28(21):4317–4331
Wamalwa BM, Zhao G, Sakka M, Shiundu PM, Kimura T, Sakka K (2007) High-level heterologous expression of Bacillus halodurans putative xylanase xyn11a (BH0899) in Kluyveromyces lactis. Biosci Biotechnol Biochem 71(3):688–693
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227(5259):680–685
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31(3):426–428
Andre L de SQ, Jorge LCC, Elza Fernandes dA, Virginia Maria CA (2006) Partial purification and characterization of xylanase produced by penicillium expansum. Brazilian Arch Biol Technol 49(3):475–480
Madden T (2013) The BLAST sequence analysis tool. In: The NCBL handbook. NCBI, Bethesda
Rognes T, Flouri T, Nichols B, Quince C, Mahé F (2016) VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584
Capra JA, Singh M (2007) Predicting functionally important residues from sequence conservation. Bioinformatics 23(15):1875–1882. https://doi.org/10.1093/bioinformatics/btm270
Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y (2015) The I-TASSER Suite: protein structure and function prediction. Nat Methods 12(1):7–8
Sánchez IE, Beltrao P, Stricher F, Schymkowitz J, Ferkinghoff-Borg J, Rousseau F, Serrano L (2008) Genome-wide prediction of SH2 domain targets using structural information and the FoldX algorithm. PLoS Comput Biol 4(4):e1000052
Schymkowitz JW, Rousseau F, Martins IC, Ferkinghoff-Borg J, Stricher F, Serrano L (2005) Prediction of water and metal binding sites and their affinities by using the Fold-X force field. Proc Natl Acad Sci USA 102(29):10147–10152
Kiel C, Wohlgemuth S, Rousseau F, Schymkowitz J, Ferkinghoff-Borg J, Wittinghofer F, Serrano L (2005) Recognizing and defining true Ras binding domains II: in silico prediction based on homology modelling and energy calculations. J Mol Biol 348(3):759–775
Senthilkumar B, Meshachpaul D, Sethumadhavan R, Rajasekaran R (2015) Selection of effective and highly thermostable Bacillus subtilis lipase a template as an industrial biocatalyst-A modern computational approach. Front Biol 10(6):508–519
Pedretti A, Villa L, Vistoli G (2004) VEGA–an open platform to develop chemo-bio-informatics applications, using plug-in architecture and script programming. J Computer-Aided Mol Des 18(3):167–173
Pearson WR (2013) An introduction to sequence similarity (“homology”) searching. Curr Protoc Bioinform. https://doi.org/10.1002/0471250953.bi0301s42
Lin JH (1991) Divergence measures based on the shannon entropy. Ieee Trans Inform Theory 37(1):145–151. https://doi.org/10.1109/18.61115
Lobanov MY, Bogatyreva NS, Galzitskaya OV (2008) Radius of gyration as an indicator of protein structure compactness. Mol Biol 42(4):623–628. https://doi.org/10.1134/S0026893308040195
Momen-Roknabadi A, Sadeghi M, Pezeshk H, Marashi SA (2008) Impact of residue accessible surface area on the prediction of protein secondary structures. BMC Bioinformatics 9(1):357. https://doi.org/10.1186/1471-2105-9-357
Zhang W, Mullaney EJ, Lei XG (2007) Adopting selected hydrogen bonding and ionic interactions from Aspergillus fumigatus phytase structure improves the thermostability of Aspergillus niger PhyA phytase. Appl Environ Microbiol 73:3069–3076
Ohmura T, Ueda T, Hashimoto Y, Imoto T (2001) Tolerance of point substitution of methionine for isoleucine in hen egg white lysozyme. Protein Eng 14(6):421–425
Acknowledgements
We are thankful to Higher Education commission (HEC) Pakistan for providing the funding for this research project [Grant No. PM-IPFP/HRD/HEC/2011/0018] and scholarship funding for HEC Indigenous Ph D Fellowship 5000 Phase II” Award No. 315-2158-2BS3-029.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mahmood, M.S., Rasul, F., Saleem, M. et al. Characterization of recombinant endo-1,4-β-xylanase of Bacillus halodurans C-125 and rational identification of hot spot amino acid residues responsible for enhancing thermostability by an in-silico approach. Mol Biol Rep 46, 3651–3662 (2019). https://doi.org/10.1007/s11033-019-04751-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-019-04751-5