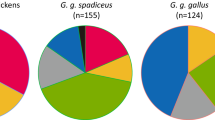Abstract
The domestic chicken (Gallus gallus domesticus) is an excellent model for genetic studies of phenotypic diversity. The Guangxi Region of China possesses several native chicken breeds displaying a broad range of phenotypes well adapted to the extreme hot-and-wet environments in the region. We thus evaluated the genetic diversity and relationships among six native chicken populations of the Guangxi region and also evaluated two commercial breeds (Arbor Acres and Roman chickens). We analyzed the sequences of the D-loop region of the mitochondrial DNA (mtDNA) and 18 microsatellite loci of 280 blood samples from six Guangxi native chicken breeds and from Arbor Acres and Roman chickens, and used the neighbor-joining method to construct the phylogenetic tree of these eight breeds. Our results showed that the genetic diversity of Guangxi native breeds was relatively rich. The phylogenetic tree using the unweighed pair-group method with arithmetic means (UPGAM) on microsatellite marks revealed two main clusters. Arbor Acres chicken and Roman chicken were in one cluster, while the Guangxi breeds were in the other cluster. Moreover, the UPGAM tree of Guangxi native breeds based on microsatellite loci was more consistent with the genesis, breeding history, differentiation and location than the mtDNA D-loop region. STRUCTURE analysis further confirmed the genetic structure of Guangxi native breeds in the Neighbor-Net dendrogram. The nomenclature of mtDNA sequence polymorphisms suggests that the Guangxi native chickens are distributed across four clades, but most of them are clustered in two main clades (B and E), with the other haplotypes within the clades A and C. The Guangxi native breeds revealed abundant genetic diversity not only on microsatellite loci but also on mtDNA D-loop region, and contained multiple maternal lineages, including one from China and another from Europe or the Middle East.





Similar content being viewed by others
References
FAO, Rischkowsky B, Pilling D (eds) (2007) The state of the world’s animal genetic resources for food and agriculture. Food and Agriculture Organization of the United Nations, Rome
Liu YP, Wu GS, Yao YG, Miao YW, Luikart G, Baig M, Beja-Pereira A, Ding ZL, Palanichamy MG, Zhang YP (2006) Multiple maternal origins of chickens: out of the Asian jungles. Mol Phylogenet Evol 38:12–19. doi:10.1016/j.ympev.2005.09.014
Miao YW, Peng MS, Wu GS, Ouyang YN, Yang ZY, Yu N, Liang JP, Pianchou G, Beja-Pereira A, Mitra B, Palanichamy MG, Baig M, Chaudhuri TK, Shen YY, Kong QP, Murphy RW, Yao YG, Zhang YP (2013) Chicken domestication: an updated perspective based on mitochondrial genomes. Heredity 110:277–282. doi:10.1038/hdy.2012.83
Zhao SG (2006) Genetic diversity of mtDNA sequences in Chinese indigenous chickens. Dissertation, Gansu Agricultural University (In Chinese)
Tadano R, Sekino M, Nishibori M, Tsudzuki M (2007) Microsatellite marker analysis for the genetic relationships among Japanese long-tailed chicken breeds. Poult Sci 86:460–469. doi:10.1093/ps/86.3.460
Groeneveld LF, Lenstra JA, Eding H, Toro MA, Scherf B, Pilling D, Negrini R, Finlay EK, Jianlin H, Groeneveld E, Weigend S (2010) Genetic diversity in farm animals—a review. Anim Genet 41:6–31. doi:10.1111/j.1365-2052.2010.02038.x
Ceccobelli S, di Lorenzo P, Lancioni H, Monteagudo Ibáñez LV, Tejedor MT, Castellini C, Landi V, Martínez Martínez A, Delgado Bermejo JV, Vega Pla JL, Leon Jurado JM, García N, Attard G, Grimal A, Stojanovic S, Kume K, Panella F, Weigend S, Lasagna E (2015) Genetic diversity and phylogeographic structure of sixteen Mediterranean chicken breeds assessed with microsatellites and mitochondrial DNA. Livest Sci 175:27–36. doi:10.1016/j.livsci.2015.03.003
Wilkinson S, Wiener P, Teverson D, Haley CS, Hocking PM (2011) Characterization of the genetic diversity, structure and admixture of British chicken breeds. Anim Genet 43:552–563. doi:10.1111/j.1365-2052.2011.02296.x
Muchadeyi FC, Eding H, Wollny CBA, Groeneveld E, Makuka SM, Shamseldin R, Simianer H, Weigend S (2007) Absence of population substructuring in Zimbabwe chicken ecotypes inferred using microsatellite analysis. Anim Genet 38:332–339. doi:10.1111/j.1365-2052.2007.01606.x
Thomson VA, Ophélie L, Austin JJ, Hunt TL, Burney DA, Denham T, Rawlence NJ, Wood JR, Gongora J, Fink LG, Linderholm A, Dobney K, Larson G, Cooper A (2014) Using ancient DNA to study the origins and dispersal of ancestral Polynesian chickens across the Pacific. Proc Nat Acad Sci USA 111:4826–4831. doi:10.1073/pnas.1320412111
FAO (2004) Guideline for development of national management of farm animal genetic diversity. Recommended microsatellite markers, available at: http://dad.fao.org
Tadano R, Sekino M, Nishibori M, Tsudzuki M (2007) Microsatellite marker analysis for the genetic relationships among Japanese long-tailed chicken breeds. Poult Sci 86:460–469
Abiye Shenkut A, Sofia M, Johansson AM (2015) Genetic diversity of five local Swedish chicken breeds detected by microsatellite markers. PLoS ONE 4:1–13. doi:10.1371/journal.pone.0120580
Qu LJ, Li XY, Xu GF, Chen KW, Yang HJ, Zhang LC, Wu GQ, Hou ZC, Xu GY, Yang N (2006) Microsatellite analysis of genetic diversity in China native chicken populations. Sci China Ser C-life Sci 36:17–26 (In Chinese)
Librado P, Rozas J (2009) DnaSP v 5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. doi:10.1093/bioinformatics/btp187
Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48. doi:10.1093/oxfordjournals.molbev.a026036
Raymond M, Rousset F (1995) An exact test for population differentiation. Evolution 49:1280–1283. doi:10.2307/2410454
Nei M, TajimaF Tateno Y (1983) Accuracy of estimated phylogenetic tree from molecular data. II. Gene frequency data. J Mol Evol 19:153–170. doi:10.1007/BF02300753
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370. doi:10.2307/2408641
Botstein D, White RL, Skolnick MH, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Human Genet 32:314–331
Marshall TC, Slate J, Kruuk LEB, Pemberton JM (1998) Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol 7:639–655. doi:10.1046/j.1365-294x.1998.00374.x
Kumar S, Tamura K, Nei M (2004) Mega 3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163. doi:10.1093/bib/5.2.150
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 7:574–578
Jakobsson M, Rosenberg N (2007) Clumpp: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 14:1801–1806. doi:10.1093/bioinformatics/btm233
Rosenberg NA (2004) Distruct: a program for the graphical display of population structure. Mol Ecol Notes 1:137–138. doi:10.1046/j.1471-8286.2003.00566.x
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of cluster of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. doi:10.1111/j.1365-294X.2005.02553.x
Bhuiyan MSA, Chen SY, Faruque S, Bhuiyan AKFH, Beja-Pereira A (2013) Genetic diversity and maternal origin of Bangladeshi chicken. Mol Biol Rep 40:4123–4128. doi:10.1007/s11033-013-2522-6
Kawaba K, Worawat R, Taura S, Shimogiri T, Nishida T, Okamoto S (2014) Genetic diversity of mtDNA D-loop polymorphisms in Laotian native fowl populations. Asian Australas J Anim Sci 27:19–23. doi:10.5713/ajas.2013.13443
Hoque MR, Choi NR, Sultana H, Kang BS, Heo KN, Hong SK, Jo CJ, Lee H (2013) Phylogenetic analysis of a privately-owned Korean native chicken population using mtDNA D-loop variations. Asian Australas J Anim 26:157–162. doi:10.5713/ajas.2012.12459
Torroni A, Schurr TG, Cabell MF, Brown MD, Neel JV, Larsen M (1993) Asian affinities and continental radiation of the four founding native american mtdnas. Am J Hum Genet 53:563–590
Ward RH, Redd A, Valencia D, Frazier B, Bo S (1993) Genetic and linguistic differentiation in the Americas. Proc Natl Acad Sci USA 22:10663–10667. doi:10.1073/pnas.90.22.10663
Pirany N, Romanov MN, Prasad DT, Ganpule SP, Devegowda G (2007) Microsatellite analysis of genetic diversity in Indian chicken populations. J Poult Sci 44:19–28. doi:10.2141/jpsa.44.19
Osei-Amponsah R, Kayang BB, Naazie A, Osei YD, Youssao IAK, Yapi-Gnaore VC, Tixier-Boichard M, Rognon X (2010) Genetic diversity of Forest and Savannah chicken populations of Ghana as estimated by microsatellite markers. Anim Sci J 81:297–303. doi:10.1111/j.1740-0929.2010.00749.x
Ohno S (1997) The one ancestor per generation rule and three other rules of mitochondrial inheritance. Proc Natl Acad Sci USA 94:8033–8035. doi:10.1073/pnas.94.15.8033
Cuc NTK, Weigend S, Tieu HV, Simianer H (2011) Conservation priorities and optimum allocation of conservation funds for Vietnamese local chicken breeds. J Anim Breed Genet 128:284–294. doi:10.1111/j.1439-0388.2010.00911.x
Acknowledgments
We would like to think Dr. Fred Bogott at the Medical Center, Austin of Minnesota for his excellent English editing of the manuscript. This work was supported by Guangxi Provincial Natural Science Foundation of China (NO. 2013GXNSFDA019013 to Dr. Yuying Liao).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liao, Y., Mo, G., Sun, J. et al. Genetic diversity of Guangxi chicken breeds assessed with microsatellites and the mitochondrial DNA D-loop region. Mol Biol Rep 43, 415–425 (2016). https://doi.org/10.1007/s11033-016-3976-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-016-3976-0