Abstract
The exploration of genetically superior accessions is the key source of germplasm conservation and potential breeding material for the future. To meet the demand of better yielding chickpea cultivars in Pakistan the present study was organized to select more stable and resistant lines from indigenous as well as exotic chickpea germplasm obtained from Plant Genetic Resource Institute (PGRI), National Agricultural Research Centre, Islamabad, Pakistan. For the identification and evaluation of chickpea wilt resistant lines against Fusarium oxysporum f. sp. ciceris (Schlechtends), the germplasm was tested in the field for the selection of wilt resistant lines and the PCR based molecular markers were investigated to use Marker Assisted Selection (MAS) for selection of the desirable cultivars. In field trial, 70 % accessions were resistant to wilt disease, while the remaining 30 % have shown susceptibility to the disease. A total of 5 RAPD and 15 SSR markers were screened for molecular based characterization of wilt response. The data of molecular markers were scored by the presence (1) and absence (0) of allele and subjected to statistical analysis. The analysis was based on coefficient of molecular similarity using UPGMA and sorted the germplasm into two groups based on disease response. Among the total used RAPD/SSR primers, only TA194 SSR marker showed linkage to wilt resistant locus at 85 % probability. The linkage of a marker was reconfirmed by receiver operating characteristic curve. The use of the sorted wilt resistant genotypes through SSR marker TA194 can make available ample prospect in MAS breeding for yield improvement of the crop in Pakistan.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Chickpea (Cicer arietinum L.) is an important food legume and a protein rich cash crop has been classified into two main types; small dark-seeded Desi type of Indian origin and large light-seeded Kabuli type of Mediterranean origin [1]. Pakistan is the major grower country of chickpea in the world, where it is cultivated on about one million hectares with a total production of 760 thousand tons [2]. While, in Khyber Pakhtunkhwa it was cultivated on an area of 42 thousand hectares with 20 thousand tons annual production [3]. Although it is grown on large area, but the main reasons of its very low yield and production are either biotic/abiotic stresses, selection strategies for development of desirable traits cultivars and poor labour management [4–8]. In addition among various environmental constraints, one of the limiting factors which directly affected the yield and causing 10–90 % loss to the crop [9], is the fungal disease caused by Fusarium oxysporum sp. ciceris (Schlechtends) which causes chickpea wilting. Chickpea wilt is gradually prevailing in Pakistan as a result of the increased drought condition since last few years. Therefore, the issue needs great attention to enhance the yield [10]. The disease is soil or seed born [11], which is difficult to control by the use of chemicals or fungicides [12]. To overcome this serious problem, the use of resistant and quality cultivars to control wilt is the best and cheapest way for breeders to adopt [13]. In Pakistan there does exist a wide gap between its potential and real yield attributed by different constraints; unfortunately in traditional farming system the farmers still in use of old chickpea cultivars and varieties due to unavailability of the attainments of chickpea upgrading research programs to increase the yield of a crop at homestead level. However for substantial increase in the efficiency of chickpea production which is the requirement of developing countries like Pakistan to overcome on food problems, needed to adopt the use of quality seeds with allied scientific technologies by the chickpea growers. Chickpea production in the country can be stabilized and improved by the development of suitable chickpea cultivars adaptable for all sorts of environments [14]. The selection and inheritance of the desirable traits is now become possible with the advancement of Marker Assisted Selection (MAS) which provides a beneficial source to exploit the potentiality of genes against agronomic traits [15, 16]. In this connection a set of PCR based currently available RAPD and SSR markers are often chosen for their higher genome coverage [17, 18]. In previous studies the linkage map of resistance genes for FOC 1–5 races was developed using different RAPD and SSR markers in recombinant inbred lines (RILS) populations generated from various resistant and susceptible parental combinations [19–22]. While at least eight races of this fungus have been reported, out of which six are more virulent causing wilt disease [23, 24]. However, there is no any information about the existence of races in Pakistan. It has also been reported by many workers [25–28], that virulent races of the pathogens need continuous characterization for screening of germplasm because of constantly changing their nature after some time from resistant to susceptible. In addition, the conventional pathotyping techniques are no more valid for reliable evaluation and identification of wilt causing fungal pathogens [29]. Therefore, the present study was organized to select the resistant and susceptible lines in unreported chickpea local (Pakistani) and exotic (USA) germplasm through a set of RAPD and SSR markers linkage to resistance genes for future resistance gene pyramiding and to enhance resistant germplasm resources for increasing yield of chickpea in Pakistan.
Materials and methods
Plant materials
Twenty-four indigenous and 46 exotic accessions of chickpea were obtained from Plant Genetic Resource Institute (PGRI), National Agriculture Research Centre, Islamabad, Pakistan (Catalogue) for field experiments performed in the research area of Malakand University, Chakdara, Khyber Pakhtunkhwa, Pakistan during 2009–2012 [30]. For planting the accessions, randomized complete block design (RCBD) suggested by Clewer and Scarisbrick [31] was used, keeping row to row distance 75 cm with row length of 5 m.
Disease screening
Chickpea germplasm was tested for wilt resistance in field against F. oxysporum f. sp. ciceris (FOC) using the isolates provided by the Department of Pathology, University of the Punjab, Pakistan. The fungal inoculum was increased by multiplying with sorghum grains. At the time of inoculation, each of the test isolate was mixed thoroughly to develop wilt sick bed, where the accessions were plotted in rows.
RAPD/SSR molecular markers
For Molecular characterization genetic linkages both RAPD and SSR primers were screened (Tables 1, 2). Five RAPD and 15 SSR primers were tested for genetic linkage. The DNA was extracted from dry seeds through a modified technique of Kang et al. [32]. Whereas, quality of the genomic DNA was ensured through agarose gel electrophoresis. The quantification was done through Spectrophotometer with accordance to the instructions provided in the literature of the instrument protocol booklet.
PCR amplification
To optimize the conditions for polymerase chain reaction (PCR) 25 µl of reaction mixture was prepared. For PCR reproducibility 2× concentrated solution of PCR master mixture (0.05 µl Taq DNA polymerase, Reaction buffer, 4 mM MgCl2 and 0.4 mM of each dNTP) was used in the reaction. Thermal cycling was optimized with denaturation temperature for 2 min at 94 °C, annealing temperature for 1 min at 55 °C and extension temperature 72 °C for 10 min. The PCR product was resolved on 2 % agarose gel in 1 × TBE buffer at 100 V. Tracking dye was mixed in PCR tube (containing mastermix) and short spinned to mix well. The PCR product was run and visualized the DNA profile under gel documentation system for the scoring of data for linkage analysis.
Data analysis
The observations were made in rates (%) of accessions showed wilting at seedling stage, flowering time and complete response till pods maturity by using the wilt incidence formula [33].
The degree of susceptibility and resistance to disease of each line was determined by using 1–9 rating scale given by [34], which scored = 1 for highly resistant, resistant = 3, moderately resistant = 5; susceptible = 7, and highly susceptible = 9. The data from electrophorogram was scored by the presence (1) and absence (0) of allele. The variation intensity was not taken in consideration, but the linkage of molecular marker with wilt was scored. On the basis of presence and absence of alleles (bands), cluster analysis of 70 lines was performed to sort the lines with response to disease status. Coefficient of similarity based on UPGMA was performed. For Pearson correlation t test (alpha ≤ 0.05) was applied using STATISTICA version 7 for Windows. The probability of molecular marker was estimated and confirmed through receiver operating characteristic (ROC) curve analysis.
Results
In field screening 70 % accessions were observed as resistant and 30 % found susceptible to Fusarium wilt (Table 3). According to disease rating scale the total germplasm was categorized into highly resistant (HR), with wilt incidence (%) 37.1, resistant (R), with wilt incidence (%) 21.4, moderately resistant or tolerant (MR) with 22.8 wilt incidence (%) and highly susceptible (SS) group for which wilt incidence (%) was 18.6 at seedling stage. On the other hand the disease response of both local and exotic accessions at reproductive to pod maturity stage, scored HR, R, moderately resistant (MR) and susceptible (SR) lines with wilt incidence (%) calculated as 21.4, 14.3, 17.1 and 28.6 % respectively (Table 4; Fig. 1). Thus the average value of wilt incidence of the resistant group (HR, R and MR) at seedling stage was 27.1 % that dropped to 17.6 % at reproductive stage. Contrary to this, the susceptible group (SS and SR) raised at reproductive to maturity stage from 18.6 to 28.6 % respectively. Results regarding resistance to wilt disease of chickpea lines at both seedling and pods maturity stage presented in the Table 5 showed significant and distinct variation at alpha ≤ 0.050.
Linkage of molecular markers
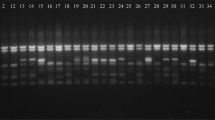
To further evaluate and identified wilt resistance lines among chickpea germplasm, five RAPD and fifteen SSR markers were investigated to assess linkage with Fusarium wilt resistance gene. These primers were selected from previous literature [35, 36]. However in present study the SSR marker TA194 has only shown significant relation with the presence of allele for resistance (Table 6); therefore, it has been selected for further analysis. The dendrogram constructed on the basis of coefficient of similarity using UPGMA divided the total germplasm into two lineages and four clusters resulted in splitting of 70 accessions into two groups. The first group displayed 77 % accessions resistant to wilt disease, while the remaining 23 % grouped as susceptible (Fig. 2). The linkage probability of TA194 marker was 85 % (Table 7), and the association of the marker was reconfirmed by ROC curve (Fig. 3). The coefficient of correlation of marker TA194 with disease resistant gene (FOC locus), Factor 1 was highly significant at P ≥ 0.01 (Table 6). The PCR amplification using TA194 however; for certain accessions have shown multiple bands (Fig. 2).
Discussion
The MAS enhance sources of distinction and make easy the complex traits selection that is otherwise time consuming process when evaluated phenotypically. The procedure of MAS for disease resistance which is typically a quantitative trait can be more efficiently developed [37]. The stability among various genotypes to select high yielding and disease free chickpea lines is the key criterion for future breeding programs. A high level of resistance in chickpea genotypes against Fusarium wilt disease has been studied [38–42]. But identification and evaluation of chickpea wilt resistant lines against F. oxysporum f. sp. ciceris aiming at to combine field screening linked with gene using PCR based markers is a new avenue in chickpea breeding in Pakistan.
The germplasm categorized on the basis of disease response at seedling and reproductive stage for comparison provided a valid conclusion and this increase in susceptibility to wilt disease was observed that may be due to slow wilting resistance of certain chickpea accessions required long time for wilting. The t-test however, indicated that chickpea both from indigenous and exotic origin showed significant variation at alpha ≤ 0.050 at seedling and reproductive stage; has already been reported [43].
For more efficient procedure to identify chickpea resistant lines in the available germplasm against Fusarium wilt disease the molecular markers can be used for chickpea screening to facilitate gene pyramiding and molecular breeding [44]. The previous workers [45], identified the genetic linkage of resistant genes using different RAPD and SSR markers for various FOC races (FOC 1, 2, 3, 4 and 5) in inbred chickpea lines developed from resistant and susceptible parental combinations. While, in our study we observed that among molecular markers (5 RAPD and 15 SSR markers) i.e., TA194 at a molecular weight 204 bp showed linkage in chickpea germplasm that was not reported earlier. Thus it was suggested that this SSR primer that successfully separated resistant (1) and susceptible lines with significant linkage to allele for resistance should be practically utilized for target chickpea breeding resistant to wilt.
The results based on dendrogram, were quite comparable with field observations. Furthermore, the linkage probability of TA194 marker was 85 %. This significant linkage of primer with resistivity against wilt disease was reconfirmed by ROC curve analysis which is recently developed for numerous agricultural applications to evaluate the performance of diagnostic experiments in the form of graphical representation [46–49].
Furthermore, in present study the coefficient of correlation of the marker TA194 with disease resistant gene (FOC locus), Factor 1 was highly significant at P ≥ 0.01. Thus the SSR marker has shown strong association with presence of allele for resistance. The PCR amplification using TA194 for certain accessions scored multiple bands, reported in earlier studies [50]. Therefore, re-synthesis of valid SSR markers is required with single amplified locus. One of the reasons of the appearance of multiple bands is the presence of cryptic sites of the primer binding sites [51]. The accessions 2273 (R) and 3058 (MR) did not show any sort of band during PCR amplification that may be due to mutation in primer binding site or absence of the locus, because these accessions were found resistant during field screening.
The evaluation and selection of superior genotypes using various scientific techniques for utilization of yield enhancement on the basis of performance stability is considered an important research study all over the world. For which the initial step is to control the devastating Fusarium wilt disease of the crop through MAS to develop disease resistant germplasm of cultivated chickpea in Pakistan. The present study however selected wilt resistant genotypes using SSR marker TA194 that can provide an opportunity in marker assisted breeding for yield improvement of the crop.
References
Khan IA, Marwat KB, Hassan G, Khan R, Ziaullah (2012) Suppressive capability of herbicides and plant extracts against chickpea weeds. J Anim Plant Sci 22:67–69
Govt. of Pakistan (2009). Economic survey of Pakistan. Ministry of Food and Agric. Federal Bureau of Statistics, pp 40
MINFAL, Agricultural Statistics of Pakistan (2009) Ministry of food, agriculture and livestock. Government of Pakistan, Islamabad
Pankaj K, Deshmukh PS, Kushwaha SR, Sunita-Kumari (2001). Indian Society of Agricultural Science. Annuals of agricultural research. Division of Plant Physiology, Indian Agricultural Research Institute, New Delhi, pp 110–112
Sharif M (2004) Wheat yield gap analysis: future options for Pakistan: a report submitted in partial fulfillment of the requirement for internship programme in Agricultural Economics B.Sc. (Hons.) Agriculture
Hassanuzzaman M, Karim MF, Fattah QA, Nahar K (2007) Yield performance of chickpea varieties following application of growth regulator. Am-Eura J Sci Res 2:117–120
Canci H, Toker C (2009) Evaluation of annual wild Cicer species for drought and heat resistance under field conditions. Genet Resource Crop Evol 56:1–6
Gaur PM, Tripathi S, Gowda CLL, Ranga Rao GV, Sharma HC, Pande S, Sharma M (2010) Chickpea seed production manual, vol 28. ICRISAT, Patancheru
Jimenez-Diaz RM, Trapero-Casas A, Cabrera de la Colina J (1989) Races of Fusarium oxysporum f. sp. ciceris infecting chickpea in Southern Spain. In: Tjamos EC, Beckman CH (eds) Vascular wilt diseases of plants. NATO ASI series, vol H28. Springer Verlag, Berlin, pp 515–520
Lines M, McMurray L, Brand J, Hobson K (2008). The influence of agronomic management on seed size attributes in Kabuli chickpea. In: 14th agronomy conference, Adelaide, South Australia, pp 21–25
Jamil FF, Sarwar M, Sarwar N, Khan JA, Zahid MH, Yousaf S, Arshad HMI, Haq I (2010) Genotyping with RAPD markers resolves phenotype diversity in the Ascochyta Blight and Fusarium wilt pathogens of chickpea in Pakistan. Pak J Bot 42(2):1369–1378
Sharma KD, Chen W, Muehlbauer FJ (2005) Genetics of chickpea resistance to Fusarium wilt and a concise set of race deferential for Fusarium oxysporum f. sp. ciceris. Plant Dis 89:385–390
Mahmood M, Athar M, Aurangzeb Ali M, Saima S, Dasti AA (2011) Analysis of genetic diversity in chickpea (Cicer arietinum L.) cultivars using random amplified polymorphic DNA (RAPD) markers. Afr J Biotechnol 10(2):140–145
Bakhshi A, Akhtar LH, Maliki SR, Massdi A, Iqbali SHM, Qureshi R (2011) Grain yield stability in chickpea (Cicer arietinum L.) across environments. Pak J Bot 43(5):2947–2951
Choudhary K, Choudhary OP, Shekhawat NS (2008) Marker assisted selection: a novel approach for crop improvement. Am-Eura J Sci Res 1(2):26–30
Allahverdipoor KH, Bahramnejad B, Amini J (2011). Selection of molecular markers associated with resistance to Fusarium wilt disease in chickpea (Cicer arietinum L.) using multivariate statistical techniques. AJCS 5(13):1801–1809
Gupta PK, Varshney RK, Sharma PC, Ramesh B (1999) Molecular markers and their applications in wheat breeding. Plant Breed 118:369–390
Gupta PK, Varshney RK (2000) The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphytica 113:163–185
Winter P, Benko-Iseppon AM, Huttel B (2000) A linkage map of chickpea (Cicer arietinum L.) genome based on recombinant inbred lines from a C. arietinum × C. reticulatum cross: localization of resistance genes for Fusarium wilt races 4 and 5. Theor Appl Genet 101:1155–1163
Sharma KD, Winter P, Kahl Muehlbauer FJ (2004) Molecular mapping of Fusarium oxysporum f. sp. ciceris race 3 resistance gene in chickpea. Theor Appl Genet 108:1243–1248
Iruela M, Castro P, Rubio J, Cubero JI, Jacinto C, Millan T, Gil J (2007) Validation of QTL for resistance to Ascochyta blight linked to resistance to Fusarium wilt race 5 in chickpea (Cicer arietinum L.). Eur J Plant Pathol 119:29–37
Gowda SJM, Radika P, Kadoo NY, Mhase LB, Gupta VS (2009) Molecular mapping of wilt resistance genes in chickpea. Mol Breed 24:177–183
Jimenez-Diaz RM, Alcala-Jimenez AR, Hervas A, Trapero-Casas JL (1993) Pathogenic variability and host resistance in the Fusarium oxysporum f. sp. ciceris/Cicer arietinum. Pathosystem. In: Proceedings of the 3rd European Seminar on Fusarium mycotoxins, taxonomy, pathogenicity and host resistance. Plant Breeding and Acclimatization Institute, Radzikov, pp 87–94
Kelly A, Alcalá-Jiménez AR, Bainbridge BW, Heale BJ, Perez-Artes E, Jiménez-Díaz RM (1994) Use of genetic fingerprinting and random amplified polymorphic DNA to characterize pathotypes of Fusarium oxysporum f. sp. ciceris infecting chickpea. Phytopathology 84:1293–1298
Porta-Puglia A (1992) Variability in Ascochyta rabiei. In: Singh KB, Saxena MC (eds) Disease resistance breeding in chickpea. Proceedings of the consultative meeting on breeding for disease resistance in Kabuli chickpea. ICARDA, Aleppo Syria, pp 135–143
Jamil FF, Sarwar M, Haq I, Bashir N (1995) Identification of pathotypes in Ascochyta rabiei (Pass) Lab., the cause of chickpea blight in Pakistan. Pak J Bot 27:193–199
Haware MP, Nene YL (1982) Races of Fusarium oxysporum f. sp. ciceri. Plant Dis 66:809–810
Jamil FF, Sarwar M, Sarwar N, Khan AJ, Zahid MH, Yousaf S, Arshid HMI, Haq I (2010) Genotyping with RAPD markers resolves pathotype diversity in the Ascochyt blight and Fusarium wilt pathogens of chickpea in Pakistan. Pak J Bot 42:1369–1378
Jamil FF, Sarwar N, Sarwar M, Khan JA, Geistlinger J, Khal G (2000) Genetic and pathogenic diversity within Ascochyta rabiei (Pass.) Lab., population in Pakistan causing blight of chickpea (Cicer arietinum L.). Physiol Mol Plant Pathol 57:243–254
Ahmad Z, Mumtaz AS, Nisar M, Khan N (2012) Diversity analysis of chickpea (Cicer arietinum L.) germplasm and its implications for conservation and crop breeding. J Agric Sci 3(5):723–731
Clewer AG, Scarisbrick DH (2001) Practical statistics and experimental designs for plant and crop science. Wiley, New York, p 331
Kang HW, Cho YG, Yoon UH, Eun MY (1998) A rapid DNA extraction method for RFLP and PCR analysis from a single dry seed. Plant Mol Biol Report 16(1):1–9
Mehrotra RS, Aggarwal A (2003) Variability in F. oxysporum f. sp. ciceris. Plant Pathol 1–2:2543
Iqbal SM, Rauf CA, Bakhsh A, Iqbal U (2005) Variability in F. oxysporum f. sp. ciceris. Mycopathologia 3(1–2):47–51
Agrawal PK, Datta S, Kumar S (2006) Pyramiding of resistance genes for Fusarium wilt in chickpea. Int Pulses Newsl 4
Datta J, Lal N (2011) Characterization of genetic diversity in Cicer arietinum L. and Cajanus cajan L. Millspaugh using random amplified polymorphic DNA and simple sequence repeat markers. Genom Quant Genet 3:30–41
Calonnec A, Wiedemann-Merdinoglu S, Deliere L, Cartolaro P, Schneider C, Delmotte F (2012) The reliability of leaf bioassays for predicting disease resistance on fruit. A case study on grapevine resistance to downy and powdery mildew. Plant Pathol. doi:10.1111/j.1365-3059.2012.02667.x
Ahmed Q, Sharma RN (1990) Source of resistance to Fusarium wilt of chickpea in Bihar. Int Chickpea Newsl 23:20
Reddy MV, Raju TN, Pundir RPO (1990) Additional source of resistance to wilt and root rots in chickpea. Int Chickpea Newsl 22:36–38
Iqbal MJ, Iftikhar K, Ilyas MB (1993) Evaluation of chickpea germplasm for resistance against wilt disease (Fusarium oxysporum). J Agric Res 31(4):449–453
Yu KH, Su T (1997) Pot screening of chickpea germplasm lines against wilt. Int Chickpea Newsl 4:19–20
Iftikhar K, Kahlid T, Ilyas MB (1997) Field screening of chickpea germplasm for the sources of resistance against Fusarium wilt. Pak J Phytopathol 9(1):31–33
Ansar M, Sheikh MI, Najma A, Yasmeen A, Abida A (2010) Identification of resistant sources in chickpea against Fusarium wilt. Pak J Bot 42(1):417–426
Soregoan CD, Ravi Kumar RL (2010) Marker Assisted characterization of wilt resistance in productive chickpea genotypes. Electron J Plant Breed 1(4):1159–1163
Sharma KD, Muehlbauer FJ (2007) Fusarium wilt of chickpea: physiological specialization genetics of resistance and resistance gene tagging. Euphytica. doi:10.1007/s10681-007-9401-y
Yuen J (2006) Deriving decision rules. Plant Health Instr. doi:10.1094/PHI-A-200517-01
Dewdney MM, Biggs AR, Turechek WW (2007) A statistical comparison of the reliability of the blossom blight forecasts of MARYBLYT and Cougarblight with receiver operating characteristic (ROC) curve analysis. Phytopathology 97:1164–1176
Wang S, Yin Y, Ma Q, Tang X, Hao D, Xu Y (2012) Genome-scale identification of cell-wall related genes in Arabidopsis based on co-expression network analysis. BMC Plant Biol 12:138
Wray NR, Yang J, Goddard ME, Visscher PM (2010) The genetic interpretation of area under the ROC curve in genomic profiling. PLoS Genet 6(2):e1000864. doi:10.1371/journal.pgen.1000864
Holton T, Christopher AJT, Mcclure L, Harker N, Henry RJ (2002) Identification and mapping of polymorphic SSR markers from expressed gene sequences of barley and wheat. Mol Breed 9:63–71
Winter P, Pfaff T, Udupa SM, Hüttel B, Sharma PC, Sahi S, Arreguin-Espinoza R, Weigand F, Muehlbauer FJ, Kahl G (1999) Characterization and mapping of sequence-tagged microsatellite sites in the chickpea (Cicer arietinum L.) genome. Mol Gen Genet 262:90–101
Acknowledgments
This study was partially supported by Pakistan Science Foundation Islamabad, Pakistan.
Conflict of interest
All the authors declared that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Ahmad, Z., Mumtaz, A.S., Ghafoor, A. et al. Marker Assisted Selection (MAS) for chickpea Fusarium oxysporum wilt resistant genotypes using PCR based molecular markers. Mol Biol Rep 41, 6755–6762 (2014). https://doi.org/10.1007/s11033-014-3561-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3561-3