Abstract
Immunoproteasomes are primarily induced upon infection and formed by replacing constitutive beta subunits with inducible beta subunits which possess specific cleavage properties that aid in the release of peptides necessary for MHC class I antigen presentation. In this study, we report the molecular characterization and expression analysis of the inducible immunosubunits PSMB8, PSMB9, PSMB9-L, and PSMB10 from rock bream, Oplegnathus fasciatus. The three subunits shared common active site residues and were placed in close proximity to fish homologues in the reconstructed phylogenetic tree, in which the mammalian homologues formed separate clades, indicating a common ancestral origin. The rock bream immunosubunits possessed higher identity and similarity with the fish homologues. RbPSMB8, RbPSMB9, RbPSMB9-L, and RbPSMB10 were multi-exonic genes with 6, 6, 7 and 8 exons, respectively. These four genes were constitutively expressed in all the examined tissues. Immunostimulants such as lipopolysaccharide and poly I:C induced RbPSMB8, RbPSMB9, RbPSMB9-L, and RbPSMB10 in liver and head kidney, suggesting their possible involvement in immune defense in rock bream.
Similar content being viewed by others
Introduction
Protein biosynthesis and degradation are two essential and highly regulated processes performed in distinct cellular compartments. Protein turnover function is performed by large inherently repressed, multisubunit, self-compartmentalizing, multicatalytic complexes called “proteasomes”. Proteasomal activity is essential to many cellular functions including DNA repair, cell cycle regulation, transcription, signal transduction, and antigen presentation [1, 2].
Proteasome architecture includes a core particle (CP) and a regulatory particle (RP). The CP (20S proteasome) is a cylindrical structure composed of four stacked rings with dyad symmetry. The outer two rings are composed of seven α-subunits (α1–α7) and the inner rings are made up of seven different β-type subunits (β1–β7). The inner surface of the interior chamber promotes protein unfolding. The α- and β-subunits share structural and sequence similarity. The α- and β-subunits maintain significant functional differences associated with their distinct N-termini. The N-terminal residues of the α-subunit form a gate at the center of the ring that restricts substrates from entering the proteasome in the absence of an activator. The β-subunit N-termini possess the proteolytic active sites. A threonine side chain is used as the attacking nucleophile and the free N-terminal amine to activate a water molecule that is embedded into the product during hydrolysis. Among the seven β-subunits, only three (β1, β2, β5; also referred to as Δ, Z and X) possess proteolytic sites [3, 4].
Proteolytic catalytic activity is exerted by the central core (20S proteasome). The inherently restrained 20S proteasome associates with different families of activators or regulators forming the mature 26S proteasome and opening access to the central proteolytic chamber. The first step in the degradation pathway is labeling of the proteins with ubiquitin molecules followed by degradation of the marked proteins by the mature 26S proteasome [4].
The CP performs three types of catalytic activities in the interior chamber including caspase-like, trypsin-like and chymotrypsin-like activities, provided by β5, β2, and β1- subunits, respectively. β5, β2, and β1- subunits possess preferential cleavage after acidic, basic, and hydrophobic amino acid residues, respectively. During an immune response, upon regulatory induction by inflammatory cytokines such as interferon gamma (IFNγ), the constitutively expressed β-subunits possessing the proteolytic sites (β1, β2, and β5) are replaced by three similar catalytic β-counterparts known as immunosubunits (β1i, β2i, β5i), and form the “immunoproteasome” (IP). In vertebrates, each catalytic subunit is encoded by two genes; one set constitutively expressed in all cell types, whereas the other set is encoded by immunosubunits coordinately expressed in immune cells such as antigen presenting cells and dendritic cells [5].
The incorporation of immunosubunits into the immunoproteasome (which possesses enhanced chymotrypsin-like, trypsin-like activities and reduced caspase-like activity), induces altered proteolytic characteristics that are favorable for antigen processing and efficient release of MHC class I ligands [6]. The peptides produced by proteolytic cleavage are translocated into the endoplasmic reticulum (ER) through transporters associated with antigen processing. In the ER, the peptides assemble with the newly synthesized MHC class I molecules and are transported to the cell surface, where they are recognized by cytotoxic T lymphocytes [7, 8]. Thus, the proteasomes play a pivotal role in the adaptive immune system [9, 10]. Multi-catalytic endopeptidase complex-like 1 (MECL1, β2i, proteasome [prosome, macropain] subunit, beta type 10 [PSMB10]) requires low molecular weight protein-2 (ip-LMP2, β1i, or PSMB9) for efficient incorporation into proteasomes, and the pre-proteasomes containing LMP2 and MECL1 require low molecular weight protein-7 (ip-LMP7, β5i, PSMB8) for maturation and interdependent IP assembly [11, 12].
Rock bream is an economically valuable fish species in Korea, and the rock bream aquaculture industry provides income for farmers. Despite the precautions taken to sustain rock bream in a disease-free state, they are affected by pathogens. It is essential to understand the underlying basic immune mechanisms to develop novel therapeutic targets for these pathogens. In this study, we identified and characterized the inducible 20S core immunosubunits PSMB8, PSMB9, and PSMB10, designated as RbPSMB8, RbPSMB9, and RbPSMB10 at the molecular level in rock bream, and analyzed their expression post-immune challenge in vivo. A PSMB9-like gene, which is characteristic of the teleosts, identified from rock bream was termed as RbPSMB9-L and analyzed.
Materials and methods
cDNA library and gene identification
A cDNA GS-FLX shotgun library was created using the Roche’s GS-FLX titanium system (DNA Link, Republic of Korea) as described previously [13]. Three cDNA clones, which were homologous to the earlier defined proteasome cluster sequences, were rescued from the cDNA library, and confirmed by homology screening by BLAST (http://blast.ncbi.nlm.nih.gov/Blast). They were designated RbPSMB8, RbPSMB9, RbPSMB9-L and RbPSMB10.
Bacterial artificial chromosome (BAC) library construction and identification of RbPSMB8, RbPSMB9, RbPSMB9-L and RbPSMB10
A rock bream BAC library was custom constructed (Lucigen, Middleton, WI, USA) and genomic sequences of RbPSMB8, RbPSMB9, RbPSMB9-L and RbPSMB10 were identified as described previously [14, 15].
Molecular characterization of RbPSMB8, RbPSMB9, RbPSMB9-L and RbPSMB10
The RbPSMB cDNA clones identified by BLAST were subjected to DNAssist (version 2.2) to obtain the open reading frame (ORF) and amino acid sequences [16]. The protein sequence was subjected to BLASTp analysis and confirmed with the other homologous sequences available in GenBank. The conserved domains of the RbPSMB protein sequences were obtained using the CDD available in NCBI (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). Multiple sequence alignment (MSA), and pairwise alignment were performed using ClustalW version 2 [17]. The phylogenetic relationship with other PSMB8, PSMB9, PSMB9-L and PSMB10 homologs obtained from GenBank was determined using the minimum evolution method available in the MEGA 5.0 program employing 5,000 bootstrap tests [18]. The amino acid identity percentages were calculated by the MatGAT program using default parameters [19]. The mRNA and genomic sequences of other PSMB homologs used for comparison of exon–intron structures were retrieved from the exon view of the Ensembl database, and those obtained from GenBank were aligned using Spidey. Putative transcription factor binding sites (TFBS) were predicted using TFSEARCH [20].
Tissue distribution and transcriptional analysis post-immune challenge
Animal rearing and tissue collection for tissue distribution analysis
Healthy rock bream fish (mean weight, ~50 g) were obtained from the Ocean and Fisheries Research Institute (Jeju, Republic of Korea). The animals were adapted to laboratory conditions (salinity 34 ± 1 ‰, pH 7.6 ± 0.5 at 24 ± 1 °C) in 400 L tanks. Tissues of liver, brain, kidney, head kidney, spleen, intestine, muscle, and skin were harvested on ice from three healthy animals and immediately snap-frozen in liquid nitrogen and stored in −80 °C, for RNA extraction.
Lipopolysaccharide (LPS) and polyinosinic:polycytidylic acid (poly I:C) challenge
For investigating the transcriptional expression of RbPSMB8, RbPSMB9, RbPSMB9-L and RbPSMB10 post-infection, time-course experiments were performed with rock breams injected with LPS or poly I:C. Purified Escherichia coli LPS purchased from Sigma-Aldrich (055:B5) was dissolved in phosphate buffered saline (PBS) and intraperitoneally (i.p.) administered at the rate of 125 μg per fish (~50 g). For poly I:C challenge, animals were i.p. injected with a 100 μL suspension of poly I:C in PBS (1.5 μg/μL; Sigma-Aldrich).
Three fish were used at each time point for the above challenges, and PBS-injected animals were used as controls. Tissues (liver and head kidney) from the un-injected control, PBS-injected, LPS, and poly I:C-challenged animals were collected at post-injection (p.i.) time points of 3, 6, 12, 24, and 48 h.
RNA isolation and cDNA synthesis
Total RNA was isolated from the tissues using Tri Reagent (Sigma, St. Louis, MO, USA). The concentration and purity of RNA was evaluated using a UV-spectrophotometer (BioRad, Hercules, CA, USA) at 260 and 280 nm. Purified RNA was diluted to 1 μg/μL, and a sample of 2.5 μg was used to synthesize cDNA from each tissue with the PrimeScript first strand cDNA synthesis kit (TaKaRa, Shiga, Japan), following the manufacturer’s protocol. Finally, the synthesized cDNA was diluted 40-fold and stored at −20 °C for later use.
Transcriptional analysis of RbPSMB8, RbPSMB9, RbPSMB9-L and RbPSMB10
Quantitative real-time reverse transcription polymers chain reaction (Q-PCR) was performed with gene specific primers (Table S1) and cDNAs prepared from tissues isolated from un-injected, PBS-injected, and immune-challenged fish. The rock bream β-actin gene was used as the invariant housekeeping gene (accession no. FJ975145). In brief, Q-PCR was performed in a 20 μL reaction volume containing 4 μL of diluted cDNA, 10 μL of 2× SYBR Green master mix, 0.6 μL of each primer (10 pmol/μL), and 4.8 μL of PCR grade water under the following thermal cycling conditions: one cycle of 95 °C for 3 min, followed by 35 amplification cycles of 95 °C for 20 s, 58 °C for 20 s, and 72 °C for 30 s. The baseline was set automatically by the Thermal Cycler Dice Real Time System software (version 2; TaKaRa). RbPSMB8, RbPSMB9, RbPSMB9-L and RbPSMB10 expression levels relative to that of β-actin were determined by the Livak method. The relative fold-change in expression after immune challenges was obtained by comparing immune-challenged tissues to those from the PBS-injected controls (at corresponding time points). The relative expression level calculated in each tissue was compared with respective expression level in muscle for tissue distribution profiling. All data are presented in terms of relative mRNA expressed as mean ± standard deviation (SD). All experiments were performed in triplicate. Statistical analyses were performed using the two-tailed Student’s t test for expression values with the corresponding controls from the same time point. P values < 0.05 were considered significant.
Results
Molecular characterization of RbPSMB8, RbPSMB9, RbPSMB9-L and RbPSMB10
The characteristic features of RbPSMB8, RbPSMB9, RbPSMB9-L and RbPSMB10 are compiled in Table 1. Apart from the general features, RbPSMB8 possessed 39 β-subunit interaction sites or polypeptide binding sites (Fig. 1a). CDD analysis of RbPSMB9 (Fig. 1b) and RbPSMB9-L (Fig. 1c) revealed 38 and 40 β-subunit interaction sites, respectively. RbPSMB10 had 38 β-subunit interaction sites (Fig. 1d). Both RbPSMB9 and RbPSMB10 had a polyA tail 12 bp downstream of the signal. As a common feature, RbPSMBs did not possess any mRNA instability motifs. RbPSMB8, RbPSMB9, RbPSMB9-L and RbPSMB10 nucleotide sequences were submitted to GenBank under the accession numbers KC795552, KC795553, KC818235, and KC795554 respectively.
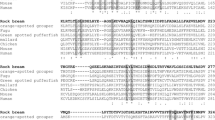
Multiple sequence alignment of RbPSMB8, RbPSMB9, RbPSMB9-L and RbPSMB10 with other homologues using ClustalW. The amino acid sequence derived from RbPSMB8 (a), RbPSMB9 (b), RbPSMB9-L (c), RbPSMB10 (d) is capitalized. The RbPSMB8, RbPSMB9, RbPSMB9-L and RbPSMB10 homologue sequences were obtained from GenBank, and the corresponding accession numbers are denoted in Tables S2, S3, S4, respectively. Identical residues are shaded and indicated by Asterisk. Highly conserved and semi-conserved residues are indicated by colon and end dot, respectively. Active site residues are indicated by Section sign, and the β-interaction sites are marked in red and underlined
The MSA revealed the conservation of all three proteins with respect to their orthologs. As expected, a high degree of conservation was observed among fish homologues. Although β-interaction sites were generally identical, variations in the degree of conservation were observed among the three genes with their respective homologues. RbPSMB8, RbPSMB9-L and RbPSMB10 shared a fairly higher preservation of residues than that of RbPSMB9 (Fig. 1a–d). A minimum evolutionary phylogenetic tree was reconstructed to understand the molecular evolution of RbPSMB8, RbPSMB9, RbPSMB9-L and RbPSMB10. The tree revealed that the rock bream PSMB proteins were placed in the fish cluster with a closer relationship with teleost homologues. The mammalian orthologues formed separate sub-clades inside each RSMB protein branch (Fig. 2). Pairwise alignment performed with the MatGAT program revealed greater identity percentages with the fish homologues (Tables S2, S3, S4). RbPSMB8 shared a similarity range of 78–98 % and an identity range of 65–95 % (Table S2). RbPSMB9 shared a similarity range of 80–98 % and an identity range of 61–94 % (Table S3). RbPSMB-9L protein shared its highest identity of 90 % with fugu and similarity range of 92–97 %. RbPSMB10 shared a similarity range of 70–97 % and an identity range of 50–90 % (Table S4).
Phylogenetic analysis of RbPSMB8, RbPSMB9, RbPSMB9-L, RbPSMB10 with other PSMB homologous sequences. The tree was constructed by the minimum evolutionary method in MEGA 5.0 using the full-length amino acids. The PSMB homologous sequences were obtained from GenBank, and the corresponding accession numbers are indicated in the Tables S2, S3, S4. Numbers above the line indicate percent bootstrap confidence values derived from 5,000 replications
Genomic characterization of RbPSMB8, RbPSMB9, RbPSMB9-L and RbPSMB10
The structural characterization of RbPSMB8 revealed the presence of six exons interrupted by five introns (Fig. 3a). The coding exon sizes were similar to that of PSMB8 in stickleback and tilapia. Although the size of the coding region in the first exon of RbPSMB8 was larger than that of Tetraodon and zebrafish, the remaining coding exons were similar in size. Additionally, RbPSMB8 showed difference in the size of the coding region with respect to the first two exons of mammalian homologues. However, high similarity was observed in the other exons. Similar to RbPSMB8, RbPSMB9 possessed six exons separated by five introns, as found in stickleback and tilapia. (Fig. 3b). RbPSMB9 shared structural similarity with the mammalian and zebrafish counterparts, with the coding region in the first exon being an exception. RbPSMB9-L gene revealed seven exon-six intron organization, unlike the medaka PSMB9-like gene (which had six exon-five intron organization). RbPSMB10 possessed eight exons separated by seven introns and its structure was similar to that of tilapia and zebrafish (Fig. 3c). RbPSMB10 shared a high homology in structure with mammalian PSMB10 homologues, with little variation in the sizes of the coding region in the first exon. RbPSMB9 and RbPSMB9-L were found to be located following the transporter-associated with antigen processing 2 (TAP2), whilst RbPSMB10 and RbPSMB8 were positioned in an opposite orientation. However, as the members of immunoproteasome subunit family, these four RbPSMBs were found to be arranged as a cluster in the PSMB locus of MHC class I region (Fig. 3d).
Genomic structural characterization of RbPSMB8, RbPSMB9 (b), RbPSMB9-L (c), RbPSMB10 (d). The genomic structures of the PSMB8, PSMB9 and PSMB10 homologues were obtained from the exon view available in the Ensembl database, and for the sequences obtained from GenBank; the structures were determined by aligning the mRNA with the genomic sequence using Spidey. The accession numbers of the homologues are indicated in brackets. a RbPSMB8, b RbPSMB9 and RbPSMB9-L, and c RbPSMB10. d Genomic organization of the immunosubunit cluster in rock bream consisted of PSMBs and TAP2. The arrows indicate the orientation of the genes
Analysis of 5′ flanking regions (~1 kb) for the putative TFBS revealed the presence of binding sequences for a number of regulatory proteins. Cis-acting elements for several transcription factors such as activator protein-1 (AP-1), CCAAT-enhancer binding protein (C/EBP), C/EBP-α and -β, cAMP response element-binding protein (CRE-BP), signal transducer and activator of transcription-x, AML-1a, Lyf-1, hepatic nuclear factor-3b, c-Rel, heat shock factor 2, interferon regulatory factor-1, and Oct-1 were present in the analyzed region of rock bream PSMBs. A putative TATA box could also be observed (Fig. S1a–d).
Tissue-specific expression of RbPSMB8, RbPSMB9, RbPSMB9-L and RbPSMB10
For a comparative transcriptional profiling, the RbPSMB mRNAs were quantified by qPCR technique using gene-specific primers. The tissue expression analysis performed in healthy rock bream tissues revealed a constitutive expression of RbPSMB8, RbPSMB9, RbPSMB9-L and RbPSMB10 genes in all the tissues examined. RbPSMB8 was highly expressed in spleen and kidney. While RbPSMB9 and RbPSMB9-L revealed similar pattern of higher expression in intestine and liver, RbPSMB10 was robustly expressed in liver (Fig. 4). It was noteworthy that significantly higher and almost similar magnitude of relative expression for all four RbPSMBs was detected in immune tissues such as intestine, liver, head kidney, spleen and kidney suggesting their immune relevance.
Tissue distribution analysis of RbPSMB8, RbPSMB9, RbPSMB9-L and RbPSMB10. RbPSMB8, RbPSMB9, RbPSMB9-L and RbPSMB10 tissue-specific expression: Mu muscle; Br brain; Sk skin; In intestine; Lr liver; Hk head kidney; Sp spleen; Kd kidney. mRNA expression was analyzed using Q-PCR. Relative mRNA expression was calculated using the Livak method, with β-actin as the invariant control gene. Relative mRNA level was compared with muscle expression to determine tissue-specific expression fold. Data are mean values (n = 3) with error bars representing the standard deviation
Transcriptional expression post-immune challenges
LPS and poly I:C are mitogenic stimulants that induce IFN-γ production. In this study, transcriptional expression was detected in the liver and head kidney sampled from rock breams challenged with LPS and poly I:C. The induction time-points and the maximum fold of expression are presented in Table 2. The liver showed induced expression of RbPSMB8, RbPSMB9, RbPSMB9-L and RbPSMB10 at all time-points post LPS challenge (Fig. 5a), whereas up-regulation of the genes in head kidney was observed only from 3 to 24 h p.i. (Fig. 5b; Table 2). In liver, the poly I:C challenge induced RbPSMB8, RbPSMB9, at all-time points, whereas RbPSMB9-L and RbPSMB10 revealed induction from 3 to 24 h (Fig. 5c). In head kidney, a significant elevation was observed from 3 to 24 h p.i. for RbPSMB8, RbPSMB9 and RbPSMB10, while from 3 to 12 h for RbPSMB9-L (Fig. 5d; Table 2).
Expression analysis of RbPSMB8, RbPSMB9, RbPSMB9-L and RbPSMB10 after immune challenges. RbPSMB8, RbPSMB9, RbPSMB9-L and RbPSMB10 expression was analyzed post-lipopolysaccharide (LPS) challenge [liver (a) and head kidney (b)], post-polyI:C challenge [liver (c) and head kidney (d)] using Q-PCR analyses. Relative mRNA expression was calculated by the Livak method relative to PBS-injected controls with β-actin as the reference gene. Data shown with Asterisk indicates significant expression levels at P < 0.05
Discussion
Diseases are a major concern in fish aquaculture, and enhancing the adaptive immune system of fish is a potential strategy for disease prevention. A mandatory step in this is to understand the antigen presenting system in fish, which activates specific T-cell responses, through the MHC class pathway. Immunoproteasomes are involved in the antigen presentation by generating the antigenic peptides [21]. Immunoproteasomes formed by substituting catalytic β-subunits with immunosubunits exhibit differential preferences for generating diverse peptides, in contrast to normal proteasomes and facilitate an improved adaptive immune response [22, 23]. In addition to the initially discovered antigen presenting function, immunoproteasomes also play a significant role in cytokine production [24, 25] and T-cell differentiation and survival [26, 27]. In this study, we characterized four rock bream immunosubunits and their expression pattern in vivo post-immune challenges.
Molecular characterization of four RbPSMBs revealed the potential active sites and β-subunit interaction sites. The deduced immunosubunit proteins in rock bream shared similar active site residues (T, D, R, K, S, D, S, and G) at different positions, which were conserved among the respective homologs. The MSA revealed high conservation among the residues (Fig. 1a–d), and the evolutionary analysis revealed closer proximity with other fish homologs (Fig. 2). The β-subunits have been found in lower eukaryotes and archaebacteria where they are involved in the degradation of full-length proteins that are defective or destined for degradation by cellular control mechanisms [28]. Antigen presentation is an additional function imposed on proteasomes during evolution. Conservation of the primary structure, which reflects functional preservation, and closer association with the fish homologs, suggesting their common ancestral origin, together affirms the significant functional role of these rock bream immunosubunits.
Vertebrate evolution is believed to have encountered two genome duplication events that resulted in paralogous regions in the genome. In humans, one hypothesis with regard to the β-type subunit cluster is that a duplication event resulted in the formation of separate constitutive and immunosubunit gene clusters which translocated to different positions in the chromosome that are syntenic in mouse as well [29, 30]. Characterization of the immunosubunit linkage and evolution has been performed in medaka and zebrafish [31, 32]. Genomic characterization revealed similarities in the structures of the homologs obtained from fish and mammals; but, some species-dependent variation was also observed. The rock bream immunosubunits were present in a single clone identified from the BAC library. Unlike medaka PSMB9-like gene, RbPSMB9-L gene possessed seven exon-six intron organization, suggesting an intron insertion event during the evolution. PSMB9-L gene is presumed to have arisen due to a cis duplication event, particularly in teleosts [31]. Interestingly, the TAP2 (Accession No: KC818236) was found in the same clone with the same orientation as found in Japanese pufferfish [31], suggesting their presence on the same locus in the chromosome as in Japanese pufferfish (Fig. 3a). The organization of the MHC class I locus, in terms of gene order and orientation, was similar to that from Japanese pufferfish [31], medaka [33], and rainbow trout [34], suggesting that the arrangement of these class Ia of MHC genes is thoroughly conserved in teleosts. It was worthy to note that this locus is believed to constitute an evolutionary stable core, since these genes have sustained a conserved organization during the vigorous rearrangement events of teleost MHC region [33].
Transcription factors determine when genes should be turned on or off and orchestrate many processes. Hence, they will pave the way for understanding the combined control of many genes. Prediction of putative TFBS have revealed the presence of significant cis-acting elements in RbPSMBs such as C/EBP α and β, AP-1, CRE-BP, and AML-1a, which play vital roles in immune responses [35–39] (Fig. 1a–d), suggesting a role for rock bream immunosubunits in immune responses.
IPs are generally destined to generate peptides for immune-surveillance. The coordinated incorporation of the immunosubunits with standard subunits (which is under tissue-specific control) suggests that the alternative mixed proteasomes (Δ/MEC1 or LMP2/Z) may be detrimental in certain cell types. IPs are present in the retina and brain, hinting at their non-immunological role [40, 41]. In addition, IPs also possess antioxidant properties [42] and regulate tumor cell growth [43, 44]. IPs are also constitutively expressed in immune tissues which are stimulated by cytokine expression-like IFN and tumor necrosis factor-α (TNF-α). IFNγ is not essential for constitutive expression of the IPs; but, is essential for their up-regulation in mice [45]. MHC-related genes were determined to be highly expressed in lymphoid tissues in rainbow trout [34]. The investigation of the distribution of rock bream immunosubunits in tissues revealed their highest expression in immune related tissues like kidney, spleen, head kidney, liver, and intestine; whereas, moderate levels of expression were observed in skin (Fig. 4). Our results are in consistent with the transcriptional profiles of MHC-related genes in rainbow trout [34]. Spleen, kidney and head kidney are the lymphoid organs in fish [46], whereas skin and intestine are the major exposed-organs, subjected to threats by pathogenic stimulants. Hence, the significant expression of rock bream immunosubunits in these tissues is not surprising, and expected to be enhanced upon pathological conditions to play a vital role in rock bream defense.
Transcriptional induction of IPs after IFN exposure has been well demonstrated in mammals. Rock bream immunosubunit expression was analyzed by Q-PCR in fish challenged with immunostimulants such as LPS and poly I:C to elucidate whether a similar mechanism is present in fish in vivo. LPS and poly I:C stimulate IFN and TNF responses [47]. IPs are involved in antiviral humoral and innate immune responses [24], and associated with antigen presentation and processing to supply the essential peptides for T-cell responses. LMP7 helps control pathogenic immune responses, and its inhibition results in impaired cytokine production [25]. During the pathological encounter or infection, IFNγ is produced, which in turn, could increase the production of PSMBs (8, 9 and 10). Apart from IFNγ, type I IFNs (IFNα and IFNβ) also enhance IP formation [48]. In this study, rock bream immunosubunits were coordinately expressed with significant up-regulation in head kidney post LPS and poly I:C challenges, revealing their participation in antibacterial and antiviral defense in rock bream (Fig. 5b, d). The liver, enriched with macrophages and natural killer (NK) cells, is a predominant innate immune organ that plays a vital role in host defense against invading microorganisms [49–51]. An exhaustive replacement of constitutive proteasomes by IPs within 1 week was observed in mice liver during antiviral and antibacterial immune response [52] and; hence, it is no wonder that rock bream immunosubunits show an increased expression post-challenges (Fig. 5a, c). Subsequently, these immunosubunits may displace the β-type subunits which are constitutively expressed [6, 53], and thereby alter the cleavage specificity of the proteasome machinery. Finally, the modification in the subunit composition helps the antigen presentation process mediated by several proteins encoded by MHC class I region [54, 55]. Therefore, PSMBs are vital components of vertebrate immunity.
LPS is an endotoxin that stimulates immune cascade and results in the synthesis of IFNs [56–59]. Additionally, MHC class-Ib molecules present intracellular bacteria and serve as recognition elements for NK cells [60]. Recognition of poly I:C by TLR3 stimulates IFN production [61]. The coordinated up-regulation of the rock bream immunosubunits after the LPS and poly I:C challenges, which are usually employed to study the host immune responses against Gram-negative bacterial and viral infections, suggests a similar mechanism of induction as demonstrated in mammals. We previously noticed that the rock bream type I IFNs are induced in head kidney following various challenges [59]. Transcripts of rock bream type I IFNs in liver was quantified by Q-PCR. One of the IFNs was prominently induced at all the time points examined in liver post LPS-challenge compared to that of the other (Fig. S2a). IFNs are primarily induced upon viral challenge, which could be the reason why RbIFNs showed higher expression after the poly I:C challenge in liver and head kidney compared to that of the LPS [59]. These transcriptional modulations between RbIFNs and RbPSMBs suggest that there might be a functional relevance for IFN-mediated transcriptional regulation of PSMB, which require further experimental evidence.
Immunoproteasomes preserve the protein homeostasis during oxidative stress [42]. As IPs also play a major role in preventing excessive cell damage by protein turnover [62], apart from their antigen presenting function, the induction of immunosubunits after a challenge may be attributed to efficient clearing of damaged proteins resulted from consequences of oxidative stress; and thus, maintain the homeostasis and sustain the cell viability.
Proteasome subunits have been identified in Paralichthys olivaceus [63], Oryzias latipes [33], Fugu rubripes [31] and Danio rerio [32], and many of these studies have focused on gene linkage analysis. Moreover, limited reports are available on the genomic structures and expression analysis of the immunoproteasome cluster post in vivo immune challenges. In this study, we identified, characterized the rock bream immunosubunits, and investigated their transcriptional expression to develop a comparative understanding. We hope that these results will illuminate similar mechanisms of IP expression in fish as in mammals.
Conclusion
We identified and characterized the immunoproteasome genes from rock bream at the molecular level. Genomic characterization of the rock bream immunosubunits revealed the conserved exon–intron structure and further support that the order and orientation of MHC class I genes are conserved in teleosts. The protein homology was also high among different animal groups, together suggesting their functional conservation in vertebrates. Their ubiquitous mRNA expression and up-regulation post-mitogenic challenges in the immune tissues further provided evidence for their involvement in immune defense of rock bream.
References
Pickart CM, Cohen RE (2004) Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol 5(3):177–187. doi:10.1038/nrm1336
Finley D (2009) Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem 78:477–513. doi:10.1146/annurev.biochem.78.081507.101607
Xie Y (2010) Structure, assembly and homeostatic regulation of the 26S proteasome. J Mol Cell Biol 2(6):308–317. doi:10.1093/jmcb/mjq030
Kim HM, Yu Y, Cheng Y (2011) Structure characterization of the 26S proteasome. Biochim Biophys Acta 1809(2):67–79. doi:10.1016/j.bbagrm.2010.08.008
Macagno A, Gilliet M, Sallusto F, Lanzavecchia A, Nestle FO, Groettrup M (1999) Dendritic cells up-regulate immunoproteasomes and the proteasome regulator PA28 during maturation. Eur J Immunol 29(12):4037–4042. doi:10.1002/(SICI)1521-4141(199912)29:12<4037:AID-IMMU4037>3.0.CO;2-T
Fruh K, Gossen M, Wang K, Bujard H, Peterson PA, Yang Y (1994) Displacement of housekeeping proteasome subunits by MHC-encoded LMPs: a newly discovered mechanism for modulating the multicatalytic proteinase complex. EMBO J 13(14):3236–3244
Tanaka K (1995) Molecular biology of proteasomes. Mol Biol Rep 21(1):21–26
Murata S, Yashiroda H, Tanaka K (2009) Molecular mechanisms of proteasome assembly. Nat Rev Mol Cell Biol 10(2):104–115. http://www.nature.com/nrm/journal/v10/n2/suppinfo/nrm2630_S1.html
Monaco JJ, Nandi D (1995) The genetics of proteasomes and antigen processing. Annu Rev Genet 29:729–754. doi:10.1146/annurev.ge.29.120195.003501
Sijts EJ, Kloetzel PM (2011) The role of the proteasome in the generation of MHC class I ligands and immune responses. Cell Mol Life Sci (CMLS) 68(9):1491–1502. doi:10.1007/s00018-011-0657-y
Murata S, Udono H, Tanahashi N, Hamada N, Watanabe K, Adachi K, Yamano T, Yui K, Kobayashi N, Kasahara M, Tanaka K, Chiba T (2001) Immunoproteasome assembly and antigen presentation in mice lacking both PA28alpha and PA28beta. EMBO J 20(21):5898–5907. doi:10.1093/emboj/20.21.5898
Kisselev AF, Garcia-Calvo M, Overkleeft HS, Peterson E, Pennington MW, Ploegh HL, Thornberry NA, Goldberg AL (2003) The caspase-like sites of proteasomes, their substrate specificity, new inhibitors and substrates, and allosteric interactions with the trypsin-like sites. J Biol Chem 278(38):35869–35877. doi:10.1074/jbc.M303725200
Umasuthan N, Whang I, Lee Y, Lee S, Kim Y, Kim H, Jung SJ, Oh MJ, Choi CY, Yeo SY, Lee SJ, Lee J (2011) Heparin cofactor II (RbHCII) from rock bream (Oplegnathus fasciatus): molecular characterization, cloning and expression analysis. Fish Shellfish Immunol 30(1):194–208. doi:10.1016/j.fsi.2010.10.004
Revathy KS, Umasuthan N, Whang I, Lee Y, Lee S, Oh MJ, Jung SJ, Choi CY, Park CJ, Park HC, Lee J (2012) A novel acute phase reactant, serum amyloid A-like 1, from Oplegnathus fasciatus: genomic and molecular characterization and transcriptional expression analysis. Dev Comp Immunol. doi:10.1016/j.dci.2012.03.014
Umasuthan N, Bathige S, Revathy KS, Wickramaarachchi WD, Wan Q, Whang I, Kim E, Park M, Park H-C, Lee J (2013) A C1 inhibitor ortholog from rock bream (Oplegnathus fasciatus): Molecular perspectives of a central regulator in terms of its genomic arrangement, transcriptional profiles and anti-protease activities of recombinant peptide. Dev Comp Immunol 42(2):197–210
Patterton H-G, Graves S (2000) DNAssist: the integrated editing and analysis of molecular biology sequences in Windows. Bioinformatics 16(7):652–653. doi:10.1093/bioinformatics/16.7.652
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739. doi:10.1093/molbev/msr121
Campanella JJ, Bitincka L, Smalley J (2003) MatGAT: an application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinform 4:29. doi:10.1186/1471-2105-4-29
Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, Podkolodny NL, Kolchanov NA (1998) Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic acids research 26(1):362–367
Kloetzel PM (2001) Antigen processing by the proteasome. Nat Rev Mol Cell Biol 2(3):179–187. doi:10.1038/35056572
Eggers M, Boes-Fabian B, Ruppert T, Kloetzel PM, Koszinowski UH (1995) The cleavage preference of the proteasome governs the yield of antigenic peptides. J Exp Med 182(6):1865–1870
Kloetzel PM, Ossendorp F (2004) Proteasome and peptidase function in MHC-class-I-mediated antigen presentation. Curr Opin Immunol 16(1):76–81
Hensley SE, Zanker D, Dolan BP, David A, Hickman HD, Embry AC, Skon CN, Grebe KM, Griffin TA, Chen W, Bennink JR, Yewdell JW (2010) Unexpected role for the immunoproteasome subunit LMP2 in antiviral humoral and innate immune responses. J Immunol 184(8):4115–4122. doi:10.4049/jimmunol.0903003
Muchamuel T, Basler M, Aujay MA, Suzuki E, Kalim KW, Lauer C, Sylvain C, Ring ER, Shields J, Jiang J, Shwonek P, Parlati F, Demo SD, Bennett MK, Kirk CJ, Groettrup M (2009) A selective inhibitor of the immunoproteasome subunit LMP7 blocks cytokine production and attenuates progression of experimental arthritis. Nat Med 15(7):781–787. doi:10.1038/nm.1978
Caudill CM, Jayarapu K, Elenich L, Monaco JJ, Colbert RA, Griffin TA (2006) T cells lacking immunoproteasome subunits MECL-1 and LMP7 hyperproliferate in response to polyclonal mitogens. Journal of immunology 176(7):4075–4082
Zaiss DMW, de Graaf N, Sijts AJAM (2008) The proteasome immunosubunit multicatalytic endopeptidase complex-like 1 Is a T-Cell-intrinsic factor influencing homeostatic expansion. Infect Immun 76(3):1207–1213. doi:10.1128/iai.01134-07
S-i Yamada, J-i Niwa, Ishigaki S, Takahashi M, Ito T, Sone J, Doyu M, Sobue G (2006) Archaeal proteasomes effectively degrade aggregation-prone proteins and reduce cellular toxicities in mammalian cells. J Biol Chem 281(33):23842–23851. doi:10.1074/jbc.M601274200
Kasahara M (1999) The chromosomal duplication model of the major histocompatibility complex. Immunol Rev 167:17–32
Kasahara M, Hayashi M, Tanaka K, Inoko H, Sugaya K, Ikemura T, Ishibashi T (1996) Chromosomal localization of the proteasome Z subunit gene reveals an ancient chromosomal duplication involving the major histocompatibility complex. Proc Natl Acad Sci USA 93(17):9096–9101
Clark MS, Pontarotti P, Gilles A, Kelly A, Elgar G (2000) Identification and characterization of a beta proteasome subunit cluster in the Japanese pufferfish (Fugu rubripes). J Immunol 165(8):4446–4452
Murray BW, Sultmann H, Klein J (2000) Identification and linkage of the proteasome activator complex PA28 subunit genes in zebrafish. Scand J Immunol 51(6):571–576
Matsuo M, Asakawa S, Shimizu N, Kimura H, Nonaka M (2002) Nucleotide sequence of the MHC class I genomic region of a teleost, the medaka (Oryzias latipes). Immunogenetics 53(10–11):930–940. doi:10.1007/s00251-001-0427-3
Hansen JD, Strassburger P, Thorgaard GH, Young WP, Du Pasquier L (1999) Expression, linkage, and polymorphism of MHC-related genes in rainbow trout, oncorhynchus mykiss. J Immunol 163(2):774–786
Sytina EV, Pankratova EV (2003) Oct-1 transcription factor–plasticity and polyfunctionality. Mol Biol 37(5):755–767
Natsuka S, Akira S, Nishio Y, Hashimoto S, Sugita T, Isshiki H, Kishimoto T (1992) Macrophage differentiation-specific expression of NF-IL6, a transcription factor for interleukin-6. Blood 79(2):460–466
Greenwel P, Tanaka S, Penkov D, Zhang W, Olive M, Moll J, Vinson C, Di Liberto M, Ramirez F (2000) Tumor necrosis factor alpha inhibits type I collagen synthesis through repressive CCAAT/enhancer-binding proteins. Mol Cell Biol 20(3):912–918
Akagi T, Thoennissen NH, George A, Crooks G, Song JH, Okamoto R, Nowak D, Gombart AF, Koeffler HP (2010) In vivo deficiency of both C/EBPβ and C/EBPε results in highly defective myeloid differentiation and lack of cytokine response. PLoS One 5(11):e15419. doi:10.1371/journal.pone.0015419
Hess J, Angel P, Schorpp-Kistner M (2004) AP-1 subunits: quarrel and harmony among siblings. J Cell Sci 117(Pt 25):5965–5973. doi:10.1242/jcs.01589
Ferrington DA, Hussong SA, Roehrich H, Kapphahn RJ, Kavanaugh SM, Heuss ND, Gregerson DS (2008) Immunoproteasome responds to injury in the retina and brain. J Neurochem 106(1):158–169. doi:10.1111/j.1471-4159.2008.05345.x
Singh S, Awasthi N, Egwuagu CE, Wagner BJ (2002) Immunoproteasome expression in a nonimmune tissue, the ocular lens. Arch Biochem Biophys 405(2):147–153. doi:10.1016/s0003-9861(02)00341-7
Seifert U, Bialy LP, Ebstein F, Bech-Otschir D, Voigt A, Schroter F, Prozorovski T, Lange N, Steffen J, Rieger M, Kuckelkorn U, Aktas O, Kloetzel PM, Kruger E (2010) Immunoproteasomes preserve protein homeostasis upon interferon-induced oxidative stress. Cell 142(4):613–624. doi:10.1016/j.cell.2010.07.036
Ho YK, Bargagna-Mohan P, Wehenkel M, Mohan R, Kim KB (2007) LMP2-specific inhibitors: chemical genetic tools for proteasome biology. Chem Biol 14(4):419–430. doi:10.1016/j.chembiol.2007.03.008
Kuhn DJ, Hunsucker SA, Chen Q, Voorhees PM, Orlowski M, Orlowski RZ (2009) Targeted inhibition of the immunoproteasome is a potent strategy against models of multiple myeloma that overcomes resistance to conventional drugs and nonspecific proteasome inhibitors. Blood 113(19):4667–4676. doi:10.1182/blood-2008-07-171637
Barton LF, Cruz M, Rangwala R, Deepe GS Jr, Monaco JJ (2002) Regulation of immunoproteasome subunit expression in vivo following pathogenic fungal infection. J Immunol 169(6):3046–3052
Deivasigamani B (2007) Structure of immune organ in edible catfish, Mystus gulio. J Environ Biol/Acad Environ Biol 28(4):757–764
Reimer T, Brcic M, Schweizer M, Jungi TW (2008) poly(I:C) and LPS induce distinct IRF3 and NF-κB signaling during type-I IFN and TNF responses in human macrophages. J Leukoc Biol 83(5):1249–1257. doi:10.1189/jlb.0607412
Shin EC, Seifert U, Kato T, Rice CM, Feinstone SM, Kloetzel PM, Rehermann B (2006) Virus-induced type I IFN stimulates generation of immunoproteasomes at the site of infection. J Clin Invest 116(11):3006–3014. doi:10.1172/JCI29832
Li Z, Diehl AM (2003) Innate immunity in the liver. Curr Opin Gastroenterol 19(6):565–571
Gao B, Jeong WI, Tian Z (2008) Liver: an organ with predominant innate immunity. Hepatology 47(2):729–736. doi:10.1002/hep.22034
Parker GA, Picut CA (2005) Liver immunobiology. Toxicol Pathol 33(1):52–62. doi:10.1080/01926230590522365
Khan S, van den Broek M, Schwarz K, de Giuli R, Diener PA, Groettrup M (2001) Immunoproteasomes largely replace constitutive proteasomes during an antiviral and antibacterial immune response in the liver. J Immunol 167(12):6859–6868
Akiyama K, Yokota K, Kagawa S, Shimbara N, Tamura T, Akioka H, Nothwang H, Noda C, Tanaka K, Ichihara A (1994) cDNA cloning and interferon gamma down-regulation of proteasomal subunits X and Y. Science 265(5176):1231–1234. doi:10.1126/science.8066462
Driscoll J, Brown MG, Finley D, Monaco JJ (1993) MHC-linked LMP gene products specifically alter peptidase activities of the proteasome. Nature 365(6443):262–264. doi:10.1038/365262a0
Gaczynska M, Rock KL, Goldberg AL (1993) Gamma-interferon and expression of MHC genes regulate peptide hydrolysis by proteasomes. Nature 365(6443):264–267. doi:10.1038/365264a0
Zhang YB, Wang YL, Gui JF (2007) Identification and characterization of two homologues of interferon-stimulated gene ISG15 in crucian carp. Fish Shellfish Immunol 23(1):52–61. doi:10.1016/j.fsi.2006.09.004
Chen WQ, Xu QQ, Chang MX, Zou J, Secombes CJ, Peng KM, Nie P (2010) Molecular characterization and expression analysis of the IFN-gamma related gene (IFN-gammarel) in grass carp Ctenopharyngodon idella. Vet Immunol Immunopathol 134(3–4):199–207. doi:10.1016/j.vetimm.2009.09.007
Sieger D, Stein C, Neifer D, van der Sar AM, Leptin M (2009) The role of gamma interferon in innate immunity in the zebrafish embryo. Dis Models Mech 2(11–12):571–581. doi:10.1242/dmm.003509
Wan Q, Wicramaarachchi WD, Whang I, Lim BS, Oh MJ, Jung SJ, Kim HC, Yeo SY, Lee J (2012) Molecular cloning and functional characterization of two duplicated two-cysteine containing type I interferon genes in rock bream Oplegnathus fasciatus. Fish Shellfish Immunol 33(4):886–898. doi:10.1016/j.fsi.2012.07.018
Soloski MJ, Szperka ME, Davies A, Wooden SL (2000) Host immune response to intracellular bacteria: a role for MHC-linked class-Ib antigen-presenting molecules. Proc Soc Exp Biol Med Soc Exp Biol Med 224(4):231–239
Matsumoto M, Seya T (2008) TLR3: interferon induction by double-stranded RNA including poly(I:C). Adv Drug Deliv Rev 60(7):805–812. doi:10.1016/j.addr.2007.11.005
Opitz E, Koch A, Klingel K, Schmidt F, Prokop S, Rahnefeld A, Sauter M, Heppner FL, Volker U, Kandolf R, Kuckelkorn U, Stangl K, Kruger E, Kloetzel PM, Voigt A (2011) Impairment of immunoproteasome function by beta5i/LMP7 subunit deficiency results in severe enterovirus myocarditis. PLoS Pathog 7(9):e1002233. doi:10.1371/journal.ppat.1002233
Kim DH, Lee SM, Hong BY, Kim YT, Choi TJ (2003) Cloning and sequence analysis of cDNA for the proteasome activator PA28-beta subunit of flounder (Paralichthys olivaceus). Mol Immunol 40(9):611–616
Acknowledgments
This study was supported by a National Fisheries Research and Development Institute (RP-2014-BT-011) Grant.
Author information
Authors and Affiliations
Corresponding author
Additional information
Saranya Revathy Kasthuri and Navaneethaiyer Umasuthan have equally contributed to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kasthuri, S.R., Umasuthan, N., Whang, I. et al. Molecular characterization and expressional affirmation of the beta proteasome subunit cluster in rock bream immune defense. Mol Biol Rep 41, 5413–5427 (2014). https://doi.org/10.1007/s11033-014-3413-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3413-1