Abstract
A rapid and easy method to screen for aberrant cDNA would be a very useful diagnostic tool in genetics since a fraction of the DNA variants found affect RNA splicing. The currently used RT-PCR methods require new primer combinations to study each variant that might affect splicing. Since MLPA is routinely used to detect large genomic deletions and successfully detected exon skipping events in Duchenne muscular dystrophy in cDNA, we performed a pilot study to evaluate its value for BRCA1 cDNA. The effect of puromycin, DNase I and two different DNA cleaning protocols were tested in the RNA analysis of lymphocyte cultures. We used two samples from unrelated families with two different BRCA1 exon deletion events, two healthy unrelated controls and six samples from hereditary breast/ovarian cancer syndrome (HBOC) patients without BRCA1/2 mutations. Using RNA treated with DNase I and cleaned in a column system from puromycin-treated fractions, we were able to identify the two BRCA1 deletions. Additional HBOC patients did not show additional splice events. However, we were not able to get reproducible results. Therefore, the cDNA-MLPA technique using kit BRCA1 P002 is in our hands currently not reliable enough for routine RNA analysis and needs further optimization.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Genetic screening of the BRCA1 and BRCA2 genes is offered to families with high risk of breast and ovarian cancer. Besides clear pathogenic mutations and polymorphisms, unclassified variants (UVs) of unclear clinical relevance are found. Some of these UVs may result in aberrant splicing, by affecting the donor or acceptor splice sites, or exonic splice site enhancer (ESE) sites [1] as predicted in silico. Additionally, deep intronic variants, which are normally ignored, may also affect splicing. One example of a deep intronic pathogenic variant is the variant CDKN2A IVS2-105A>G, which causes retention of intronic sequence [2]. Another example is the mutation c.903+409T>C in the MTRR (methionine synthase reductase) gene, which activates a pseudoexon, causing a frameshift insertion that leads to a premature stop codon [3]. Experimental proof is needed to confirm the predicted changes in RNA splicing. The experiments are usually performed using RT-PCR, for which a set of specific primers targeted to the relevant cDNA region is needed for every new variant [4–7]. It is noteworthy that exon skipping is the most common alternative splice event [8]. After the report of Kesari et al. [9], who were able to detect skipping events on cDNA from the Duchenne muscular dystrophy (DMD) gene using the respective genomic multiplex ligation probe amplification (MLPA) kit, we sought to evaluate the use of a commercially available BRCA1 MLPA kit [10] for the detection of exon skipping in cDNA instead of genomic DNA. BRCA1 MLPA is a multiplex assay based on the hybridization of a large set of primers throughout the entire coding part of the BRCA1 gene. Therefore the assay should potentially also be able to detect all exon skipping events in cDNA in the presence of a variant affecting splicing, without the need to design a specific RT-PCR assay for each variant. Although these are likely rare events, using a rapid and relatively cheap assay to assess them would be valuable in a diagnostic setting to rule out their presence.
For this pilot study, samples with BRCA1 exon 13 skipping (c.4242-1643del3835) or exon 22 skipping (c.5333-36del510) [11] were selected. The study also included samples from two unrelated healthy controls and six samples from patients belonging to high risk families for which no BRCA1 or BRCA2 mutation was identified in the standard diagnostic screening. Here we show that the MLPA method was able to detect the skipping events, but it was not reproducible enough for use in clinical testing despite the optimization attempts which are here described.
Materials and methods
Cell culture
White blood cells were isolated and cultured in complete medium consisting of: RPMI 1640 supplemented with l-glutamine (Gibco) and 12.5% FCS with additional supplements and antibiotics. Lymphocyte growth was stimulated with 50 μL/mL PHA (Gibco) and 10 U/mL of IL-2 (Roche). At day 7, 4–6 h before harvesting the cells, cultures were treated with 200 μg/mL of puromycin (Sigma), to enrich for transcripts containing premature stop codons by the inhibition of NMD [12].
RNA isolation, cDNA synthesis and MLPA reaction
Total RNA was isolated using TRIzol (Invitrogen) or TRIpure (Roche) reagent. RNA samples used were either not subjected to DNase I treatment or treated with DNA-free kit (AMBION) or with DNase I treatment followed by purification in the column system RNeasy MinElute Kit (Qiagen). First-strand cDNA was obtained with Reverse Transcriptase M-MUL (Finnzymes) using random hexamers (Invitrogen) following the manufacturers’ instructions. The cDNA was amplified with the SALSA MLPA P002 probe mix (MRC-Holland) according to the manufacturer’s protocol. Fragment analysis was performed by capillary electrophoresis in an ABI PRISM 3730 automatic sequencer (Applied Biosystems).
Data analysis
The size calling and the peak areas were assessed using the Genemarker software (Softgenetics) and exported to a “.txt” file. The values of the antisense probes were extremely low compared to the sense probes, and they do not have known biological meaning. Therefore, the data was filtered to leave only the data from probes corresponding in sequence to that of sense BRCA1 mRNA. The normalization of the data was performed using a spreadsheet according to the Manual spreadsheet-based MLPA analysis instructions (available on the MRC-Holland website: www.MLPA.com). The threshold values for deletions and duplications were set to 0.75–1.25, respectively, which are also used for DNA analysis [13–16].
Results
With the SALSA MLPA P002 kit, strong signals were obtained for 21 out of 25 probes. These probes contained more than 85% nucleotides hybridizing to the exon sequence in the correct orientation. The signals for the probes with less than 85% matching exonic sequence (exons 1A, 9 and 19) or in antisense (23) were extremely weak and often not even detectable by the software. This also confirms the absence of contaminating genomic DNA in the RNA samples.
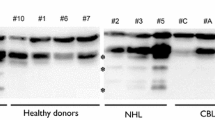
Initially, we have compared the results from puromycin-treated and non-treated samples (Fig. 1), without DNase I treatment. The results were not optimal, but it was observed that the puromycin-treated samples gave better results than the non-treated. Subsequently, we tested the effect of two different DNase I treatment options: (1) DNase I treatment followed by purification in a column system and (2) DNase I treatment kit that allows to remove the enzyme by precipitation and centrifugation. The results were considerably improved when the RNAs were cleaned in a column system (data not shown), i.e. variation in the signals among individuals was greatly reduced, at least in two independent experiments.
Six samples from high risk families without a BRCA1/2 mutation were also analyzed (data not shown) using the puromycin-treated fractions and RNAs treated with DNase I and cleaned in a column system. None of these samples showed an exon skipping event, in the 20 exons tested. However, in an independent third experiment we observed increased interindividual variability in some exon signals. Many exons had normalized values outside the 0.75–1.25 thresholds (Fig. 2). This was also observed in healthy control samples. This hampers the evaluation of splicing defects as it suggests duplications or deletions events that would need experimental follow-up or repetitive MLPA analysis to determine reproducibility.
Discussion
The MLPA method is widely used in diagnostics, mainly to test genomic events such as deletions and duplications. Although there are a few commercial RT-MLPA kits, these are designed to test the expression of genes associated with certain biological processes, MRC-Holland has not developed RT-MLPA kits to test splice events. Besides the use of the MLPA, or other multiplex approaches, to test the effect of genetic variants predicted to affect splicing at the RNA level, it would be useful to test for BRCA1 and BRCA2 mutation negative patients with strong breast and/or ovarian cancer history. This group of patients may carry variants outside the screened intronic region flanking the exons which could affect splicing. Since exon skipping is the most common alternative splice event [8], developing a test that allows to screen for exon skipping events would detect the majority of alternative splice events.
One single study has previously shown that MLPA could be used to test exon skipping events in RNA transcripts of the DMD gene [9]. Here we report the use of MLPA kit for the analysis of BRCA1 exon skipping events. The most optimal results were obtained from puromycin-treated samples and when RNA was treated with DNase I and subsequently purified in a column system. However, despite efforts to optimize the technique further, we were not able to get reliable, reproducible results for unequivocal interpretation using the kit BRCA1 P002. This variation was also observed in healthy control samples, which showed both deletion and duplication events in one out of three experiments performed.
MLPA test is a flexible multiplex assay which allows for up to a total of 50 probes and in principle, it should be possible to use it for detection of alternative splicing events other than exon skipping. To be able to test also for intron retention or insertion of pseudoexons, probes crossing over exon–exon boundaries should also be included in the assay. Although mRNA-seq technology [17] will also allow to test for aberrant splicing events in patients, MLPA could be a more cost-effective technique. However, it needs to be optimized further for routine use.
References
Cartegni L, Chew SL, Krainer AR (2002) Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 3(4):285–298
Harland M, Mistry S, Bishop DT, Newton Bishop JA (2001) A deep intronic mutation in CDKN2A is associated with disease in a subset of melanoma pedigrees. Hum Mol Genet 10(23):2679–2686. doi:10.1093/hmg/10.23.2679
Homolova K, Zavadakova P, Doktor TK, Schroeder LD, Kozich V, Andresen BS (2010) The deep intronic c.903+469T>C mutation in the MTRR gene creates an SF2/ASF binding exonic splicing enhancer, which leads to pseudoexon activation and causes the cblE type of homocystinuria. Hum Mutat 31(4):437–444. doi:10.1002/humu.21206
Bonatti F, Pepe C, Tancredi M, Lombardi G, Aretini P, Sensi E, Falaschi E, Cipollini G, Bevilacqua G, Caligo MA (2006) RNA-based analysis of BRCA1 and BRCA2 gene alterations. Cancer Genet Cytogenet 170(2):93–101
Bonnet C, Krieger S, Vezain M, Rousselin A, Tournier I, Martins A, Berthet P, Chevrier A, Dugast C, Layet V, Rossi A, Lidereau R, Frebourg T, Hardouin A, Tosi M (2008) Screening BRCA1 and BRCA2 unclassified variants for splicing mutations using reverse transcription PCR on patient RNA and an ex vivo assay based on a splicing reporter minigene. J Med Genet 45(7):438–446. doi:10.1136/jmg.2007.056895
Whiley PJ, Guidugli L, Walker LC, Healey S, Thompson BA, Lakhani SR, Da Silva LM, Investigators K, Tavtigian SV, Goldgar DE, Brown MA, Couch FJ, Spurdle AB (2011) Splicing and multifactorial analysis of intronic BRCA1 and BRCA2 sequence variants identifies clinically significant splicing aberrations up to 12 nucleotides from the intron/exon boundary. Hum Mutat 32(6):678–687. doi:10.1002/humu.21495
Brandão RD, van Roozendaal K, Tserpelis D, García EG, Blok MJ (2011) Characterisation of unclassified variants in the BRCA1/2 genes with a putative effect on splicing. Breast Cancer Res Treat 129(3):971–982. doi:10.1007/s10549-011-1599-7
Sammeth M, Foissac S, Guigó R (2008) A general definition and nomenclature for alternative splicing events. PLoS Comput Biol 4 (8). doi:10.1371/journal.pcbi.1000147
Kesari A, Pirra LN, Bremadesam L, McIntyre O, Gordon E, Dubrovsky AL, Viswanathan V, Hoffman EP (2008) Integrated DNA, cDNA, and protein studies in Becker muscular dystrophy show high exception to the reading frame rule. Hum Mutat 29(5):728–737. doi:10.1002/humu.20722
Hogervorst FBL, Nederlof PM, Gille JJP, McElgunn CJ, Grippeling M, Pruntel R, Regnerus R, van Welsem T, van Spaendonk R, Menko FH, Kluijt I, Dommering C, Verhoef S, Schouten JP, van’t Veer LJ, Pals G (2003) Large genomic deletions and duplications in the BRCA1 gene identified by a novel quantitative method. Cancer Res 63(7):1449–1453
Petrij-Bosch A, Peelen T, van Vliet M, van Eijk R, Olmer R, Drüsedau M, Hogervorst F, Hageman S, Arts P, Ligtenberg M, Meijers-Heijboer H, Klijn J, Vasen H, Cornelisse C, van ‘t Veer L, Bakker E, van Ommen G, Devilee P (1997) BRCA1 genomic deletions are major founder mutations in Dutch breast cancer patients. Nat Genet 17(4):341–345
Carter MS, Doskow J, Morris P, Li S, Nhim RP, Sandstedt S, Wilkinson MF (1995) A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem 270(48):28995–29003. doi:10.1074/jbc.270.48.28995
Barbaro M, Kotajärvi M, Harper P, Floderus Y (2011) Identification of an AluY-mediated deletion of exon 5 in the CPOX gene by MLPA analysis in patients with hereditary coproporphyria. Clin Genet:no–no. doi:10.1111/j.1399-0004.2011.01628.x
Harteveld CL, Voskamp A, Phylipsen M, Akkermans N, den Dunnen JT, White SJ, Giordano PC (2005) Nine unknown rearrangements in 16p13.3 and 11p15.4 causing α and β-thalassaemia characterised by high resolution multiplex ligation-dependent probe amplification. J Med Genet 42(12):922–931. doi:10.1136/jmg.2005.033597
Nyström A-M, Ekvall S, Thuresson A-C, Denayer E, Legius E, Kamali-Moghaddam M, Westermark B, Annerén G, Bondeson M-L (2010) Investigation of gene dosage imbalances in patients with Noonan syndrome using multiplex ligation-dependent probe amplification analysis. Euro J Med Genet 53(3):117–121
Kellander M, Riley M, Liu C Application Note: GeneMarker® Software for Multiplex Ligation-dependent Probe Amplification (MLPA™). SoftGenetics LLC
Levin J, Berger M, Adiconis X, Rogov P, Melnikov A, Fennell T, Nusbaum C, Garraway L, Gnirke A (2009) Targeted next-generation sequencing of a cancer transcriptome enhances detection of sequence variants and novel fusion transcripts. Genome Biol 10(10):R115
Acknowledgments
Rita D. Brandão was supported by grant from Fundação para a Ciência e Tecnologia (SFRH/BD/32386/2006).
Conflict of interest
No competing financial interests exist.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Brandão, R.D., Tserpelis, D., Gómez García, E. et al. Detection of exon skipping events in BRCA1 RNA using MLPA kit P002. Mol Biol Rep 39, 7429–7433 (2012). https://doi.org/10.1007/s11033-012-1575-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-012-1575-2