Abstract
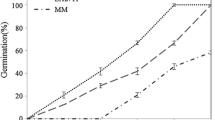
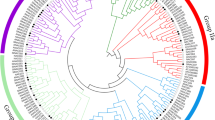
The NAC family transcription factor has demonstrated its importance in plant development and environmental stress response. Based on the microarray results under salt stress and EST information, the full-length cDNAs of two salt-inducible NAC-family genes (SlNAC1, SlNAM1) were isolated from a salt tolerant tomato cultivar, Edkawi, using Rapid Amplification of cDNA Ends (RACE). SlNAC1 and SlNAM1 encoded 301 and 296 amino acids, respectively, and the deduced protein sequences contained the typical domain of NAC-family transcription factors. Tissue expression profile analysis using semi-quantitative RT-PCR showed that SlNAC1 was expressed mainly in root, flower and green fruit; transcripts of SlNAM1 were detected in all tested tissues except for root, and high-level expression was detected in flower and matured tomato fruit. Both SlNAC1 and SlNAM1 were induced by salt stress in Edkawi, while the expression pattern was different in a salt-sensitive cultivar, ZS-5. Phylogenetic analysis for putative NAC-family peptides available in the tomato genome indicated a wide diversity of this gene family. Results obtained in the present study suggest that both SlNAC1 and SlNAM1 might play important roles in tomato stress tolerance.



Similar content being viewed by others
References
Ouyang B, Yang T, Li H, Zhang L, Zhang Y, Zhang J, Fei Z, Ye Z (2007) Identification of early salt stress response genes in tomato root by suppression subtractive hybridization and microarray analysis. J Exp Bot 58:507–520. doi:10.1093/jxb/erl258
Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9:841–857. doi:10.1105/tpc.9.6.841
Ernst HA, Olsen AN, Larsen S (2004) Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep 5:297–303. doi:10.1038/sj.embor.7400093
Sablowski RW, Meyerowitz EM (1998) A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92:93–103. doi:10.1016/S0092-8674(00)80902-2
Xie Q, Frugis G, Colgan D, Chua NH (2000) Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev 14:3024–3036. doi:10.1101/gad.852200
Ko JH, Yang SH, Park AH, Lerouxel O, Han KH (2007) ANAC012, a member of the plant-specific NAC transcription factor family, negatively regulates xylary fiber development in Arabidopsis thaliana. Plant J 50:1035–1048. doi:10.1111/j.1365-313X.2007.03109.x
Mao C, Ding W, Wu Y, Yu J, He X, Shou H, Wu P (2007) Overexpression of a NAC-domain protein promotes shoot branching in rice. New Phytol 176:288–298. doi:10.1111/j.1469-8137.2007.02177.x
Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L (2006) Overexpressing a NAM, ATAF and CUC (NAC) transcription factor enhances drought resistant and salt tolerance in rice. Proc Natl Acad Sci USA 103:12987–12992. doi:10.1073/pnas.0604882103
Bu Q, Jiang H, Li CB, Zhai Q, Zhang J, Wu X, Sun J, Xie Q, Li C (2008) Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res 18:756–767. doi:10.1038/cr.2008.53
Hu H, You J, Fang Y, Zhu X, Qi Z, Xiong L (2008) Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol 67:169–181. doi:10.1007/s11103-008-9309-5
Yokotani N, Ichikawa T, Kondou Y, Matsui M, Hirochika H, Iwabuchi M, Oda K (2009) Tolerance to various environmental stresses conferred by the salt-responsive rice gene ONAC063 in transgenic Arabidopsis. Planta 229:1065–1075. doi:10.1007/s00425-009-0895-5
Collinge M, Boller T (2001) Differential induction of two potato genes, Stprx2 and StNAC, in response to infection by Phytophthora infestans and to wounding. Plant Mol Biol 46:521–529. doi:10.1023/A:1010639225091
Oh SK, Lee S, Yu SH, Choi D (2005) Expression of a novel NAC domain-containing transcription factor (CaNAC1) is preferentially associated with incompatible interactions between chili pepper and pathogens. Planta 222:876–887. doi:10.1007/s00425-005-0030-1
Selth LA, Dogra SC, Rasheed MS, Healy H, Randles JW, Rezaian MA (2005) A NAC domain protein interacts with tomato leaf curl virus replication accessory protein and enhances viral replication. Plant Cell 17:311–325. doi:10.1105/tpc.104.027235
Fulton TM, Chunwongse J, Tanksley SD (1995) Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Mol Biol Rep 13:207–209. doi:10.1007/BF02670897
Nicot N, Hausman JF, Hoffmann L, Evers D (2005) Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56:2907–2914. doi:10.1093/jxb/eri285
Bjellqvist B, Hughes GJ, Pasquali C, Paquet N, Ravier F, Sanchez JC, Frutiger S, Hochstrasser D (1993) The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis 14:1023–1031. doi:10.1002/elps.11501401163
Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche BA, de Castro E, Lachaize C, Langendijk-Genevaux PS, Sigrist CJ (2008) The 20 years of PROSITE. Nucleic Acids Res 36:D245–D249. doi:10.1093/nar/gkm977
Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35:W585–W587. doi:10.1093/nar/gkm259
John I, Hackett R, Cooper W, Drake R, Farrell A, Grierson D (1997) Cloning and characterization of tomato leaf senescence-related cDNAs. Plant Mol Biol 33:641–651. doi:10.1023/A:1005746831643
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi:10.1007/978-1-4020-6754-9_3188
Kumar S, Nei M, Dudley J, Tamura K (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9:299–306. doi:10.1093/bib/bbn017
Ohnishi T, Sugahara S, Yamada T (2005) OsNAC6, a member of the NAC Gene family, is induced by various stresses in rice. Genes Genet Syst 80:135–139. doi:10.1266/ggs.80.135
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327. doi:10.1093/nar/30.1.325
Hegedus D, Yu M, Baldwin D, Gruber M, Sharpe A, Parkin I, Whitwill S, Lydiate D (2003) Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Mol Biol 53:383–397. doi:10.1023/B:PLAN.0000006944.61384.11
Wu Y, Deng Z, Lai J, Zhang Y, Yang C, Yin B, Zhao Q, Zhang L, Li Y, Yang C, Xie Q (2009) Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res 19:1279–1290. doi:10.1038/cr.2009.108
Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, Hayashizaki Y, Suzuki K, Kojima K, Takahara Y, Yamamoto K, Kikuchi S (2003) Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 10:239–247. doi:10.1093/dnares/10.6.239
Acknowledgments
This work was supported by the Natural Science Foundation of China (No. 30400299 and No. 30771461).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11033_2010_177_MOESM1_ESM.jpg
Gene structure of SlNAC1 (a) and SlNAM1 (b). Exons are represented as black boxes. Numbers in boxes indicate exon size in base pairs (bp). Introns are shown as lines linking exons and their sizes are shown in base pairs. The start codon is shown as ATG, and stop codon is TAA for SlNAC1 and TGA for SlNAM1. NAC or NAM domain is shown as line or dot line below the exons, and numbers on the left indicate amino acid positions of the protein domains (JPG 550 kb)
Rights and permissions
About this article
Cite this article
Yang, R., Deng, C., Ouyang, B. et al. Molecular analysis of two salt-responsive NAC-family genes and their expression analysis in tomato. Mol Biol Rep 38, 857–863 (2011). https://doi.org/10.1007/s11033-010-0177-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-010-0177-0