Abstract
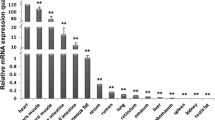
CREB3L4 (cAMP response element binding protein3 like 4) belongs to mammalian CREB/ATF subfamily transcription factors featuring a unique putative transmembrane domain lined to the bZIP region in their C-terminals. We report here the cloning of porcine CREB3L4 and its alternative splicing variant. The open reading frame of normal CREB3L4 covers 1,188 bp and encodes a 395-amino acid polypeptide, whereas its variant encodes a C-terminal truncated protein comprising only 271 amino acids. The genomic sequence of porcine CREB3L4 spans approximately 6 kb consisted of ten exons and nine introns, and maps closely to Jumping translocation breakpoint gene in a head-to-head manner on the q arm of Sus Scrofa chromosome 4. Tissue expression analysis by RT-PCR indicates that normal porcine CREB3L4 is ubiquitously expressed with relatively high abundance in stomach, liver, cerebellum, and is a predominant form in various tissues, whereas the splicing variant is expressed at extremely low level in all examined tissues. Our study will lay the groundwork for the further investigation on the physiological function of CREB3L4 in pig models.






Similar content being viewed by others
Abbreviations
- ATF:
-
Active transcription factor
- bZIP:
-
Basic region and leucine zipper
- CREB:
-
cAMP response element binding protein
- ER:
-
Endoplasmic reticulum
- HCC:
-
Hepatocellular carcinomas
- JTB:
-
Jumping translocation breakpoint
- NCBI:
-
National Center for Biotechnology Information
- S1P:
-
Site 1 protease
- TM:
-
Transmembrane
References
Garcia GE, Nicole A, Bhaskaran S, Gupta A, Kyprianou N, Kumar AP (2006) Akt-and CREB-mediated prostate cancer cell proliferation inhibition by Nexrutine, a phellodendron amurense extract. Neoplasia 8:523–533. doi:10.1593/neo.05745
Perry C, Sklan EH, Soreq H (2004) CREB regulates AChE-R-induced proliferation of human glioblastoma cells. Neoplasia 6:279–286. doi:10.1593/neo.03424
Ionescu AM, Schwarz EM, Vinson C, Puzas JE, Rosier R, Reynolds PR, O’Keefe RJ (2001) PTHrP modulates chondrocyte differentiation through AP-1 and CREB signaling. J Biol Chem 276:11639–11647. doi:10.1074/jbc.M006564200
Kim SW, Hong JS, Ryu SH, Chung WC, Yoon JH, Koo JS (2007) Regulation of mucin gene expression by CREB via a nonclassical retinoic acid signaling pathway. Mol Cell Biol 27:6933–6947. doi:10.1128/MCB.02385-06
Persengiev SP, Devireddy LR, Green MR (2002) Inhibition of apoptosis by ATFx: a novel role for a member of the ATF/CREB family of mammalian bZIP transcription factors. Genes Dev 16:1806–1814. doi:10.1101/gad.992202
Shaywitz AJ, Greenberg ME (1999) CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem 68:821–861. doi:10.1146/annurev.biochem.68.1.821
Ahn S, Olive M, Aggarwal S, Krylov D, Ginty DD, Vinson C (1998) A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol Cell Biol 18:967–977
Jia S, Noma K, Grewal SI (2004) RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science 304:1971–1976. doi:10.1126/science.1099035
Labrie C, Lessard J, Aicha SB, Savard MP, Pelletier M, Fournier A, Lavergne E, Calvo E (2008) Androgen-regulated transcription factor AIbZIP in prostate cancer. J Steroid Biochem Mol Biol 108:237–244
Stirling J, O’Hare P (2006) CREB4, a transmembrane bZip transcription factor and potential new substrate for regulation and cleavage by S1P. Mol Biol Cell 17:413–426. doi:10.1091/mbc.E05-06-0500
Qi H, Fillion C, Labrie Y, Grenier J, Fournier A, Berger L, El-Alfy M, Labrie C (2002) AIbZIP, a novel bZIP gene located on chromosome 1q21.3 that is highly expressed in prostate tumors and of which the expression is up-regulated by androgens in LNCaP human prostate cancer cells. Cancer Res 62:721–733
Levesque MH, El-Alfy M, Berger L, Labrie F, Labrie C (2007) Evaluation of AIbZIP and Cdc47 as markers for human prostatic diseases. Urology 69:196–201. doi:10.1016/j.urology.2006.11.001
Inagaki Y, Yasui K, Endo M, Nakajima T, Zen K, Tsuji K, Minami M, Tanaka S, Taniwaki M, Itoh Y, Arii S, Okanoue T (2008) CREB3L4, INTS3, and SNAPAP are targets for the 1q21 amplicon frequently detected in hepatocellular carcinoma. Cancer Genet Cytogenet 180:30–36. doi:10.1016/j.cancergencyto.2007.09.013
Stelzer G, Don J (2002) Atce1: a novel mouse cyclic adenosine 3′,5′-monophosphate-respo-nsive element-binding protein-like gene exclusively expressed in postmeiotic spermatids. Endocrinology 143:1578–1588. doi:10.1210/en.143.5.1578
Adham IM, Eck TJ, Mierau K, Muller N, Sallam MA, Paprotta I, Schubert S, Hoyer-Fender S, Engel W (2005) Reduction of spermatogenesis but not fertility in Creb3l4-deficient mice. Mol Cell Biol 25:7657–7664. doi:10.1128/MCB.25.17.7657-7664.2005
El-Alfy M, Azzi L, Lessard J, Lavergne E, Pelletier M, Labrie C (2006) Stage-specific expression of the Atce1/Tisp40alpha isoform of CREB3L4 in mouse spermatids. J Androl 27:686–694. doi:10.2164/jandrol.106.000596
Yerle M, Echard G, Robic A, Mairal A, Dubut-Fontana C, Riquet J, Pinton P, Milan D, Lahbib-Mansais Y, Gellin J (1996) A somatic cell hybrid panel for pig regional gene mapping characterized by molecular cytogenetics. Cytogenet Cell Genet 73:194–202. doi:10.1159/000134338
Cao G, Ni X, Jiang M, Ma Y, Cheng H, Guo L, Ji C, Gu S, Xie Y, Mao Y (2002) Molecular cloning and characterization of a novel human cAMP response element-binding (CREB) gene (CREB4). J Hum Genet 47:373–376. doi:10.1007/s100380200053
Hatakeyama S, Osawa M, Omine M, Ishikawa F (1999) JTB: a novel membrane protein gene at 1q21 rearranged in a jumping translocation. Oncogene 18:2085–2090. doi:10.1038/sj.onc.1202510
Wong N, Chan A, Lee SW, Lam E, To KF, Lai PB, Li XN, Liew CT, Johnson PJ (2003) Positional mapping for amplified DNA sequences on 1q21–q22 in hepatocellular carcinoma indicates candidate genes over-expression. J Hepatol 38:298–306. doi:10.1016/S0168-8278(02)00412-9
Moller M, Berg F, Riquet J, Pomp D, Archibald A, Anderson S, Feve K, Zhang Y, Rothschild M, Milan D, Andersson L, Tuggle CK (2004) High-resolution comparative mapping of pig chromosome 4, emphasizing the FAT1 region. Mamm Genome 15:717–731. doi:10.1007/s00335-004-2366-4
Nagamori I, Yabuta N, Fujii T, Tanaka H, Yomogida K, Nishimune Y, Nojima H (2005) Tisp40, a spermatid specific bZip transcription factor, functions by binding to the unfolded protein response element via the Rip pathway. Genes Cells 10:575–594. doi:10.1111/j.1365-2443.2005.00860.x
Chin KT, Zhou HJ, Wong CM, Lee JM, Chan CP, Qiang BQ, Yuan JG, Ng IO, Jin DY (2005) The liver-enriched transcription factor CREB-H is a growth suppressor protein underexpressed in hepatocellular carcinoma. Nucleic Acids Res 33:1859–1873. doi:10.1093/nar/gki332
Meyer TE, Habener JF (1993) Cyclic adenosine 3′, 5′-monophosphate response element binding protein (CREB) and related transcription-activating deoxyribonucleic acid-binding proteins. Endocr Rev 14:269–290. doi:10.1210/er.14.3.269
Yen J, Wisdom RM, Tratner I, Verma IM (1991) An alternative spliced form of FosB is a negative regulator of transcriptional activation and transformation by Fos proteins. Proc Natl Acad Sci USA 88:5077–5081. doi:10.1073/pnas.88.12.5077
Cunha AC, Weigle B, Kiessling A, Bachmann M, Rieber EP (2006) Tissue-specificity of prostate specific antigens: comparative analysis of transcript levels in prostate and non-prostatic tissues. Cancer Lett 236:229–238. doi:10.1016/j.canlet.2005.05.021
Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H (2005) Function of alternative splicing. Gene 344:1–20. doi:10.1016/j.gene.2004.10.022
Acknowledgments
We are grateful to Dr. Martine Yerle and Denis Milan (INRA, Castanet-Tolosan, France) for kindly providing the RH panel. This work was supported by the grants from 973 Programme (2006CB102100), 863 Programme (2008AA10Z134, 2006AA10Z140), High Education Doctorial Subject Research Programme (20060504016), General Programme (30771585,30600438) and Key Programe (30330440) of National Natural Science Foundation of China.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Qi, Y.M., Lei, T., Zhou, L. et al. Genomic organization, alternative splicing and tissues expression of porcine CREB3L4 gene. Mol Biol Rep 36, 1881–1888 (2009). https://doi.org/10.1007/s11033-008-9394-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-008-9394-1