Abstract
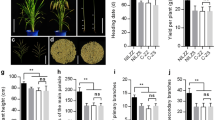
The naturally occurring genetic variation in the universal flowering (or heading date in crops) pathway has produced major advancements in crop domestication and expansion, and the various combinations of heading date genes have facilitated the plants to heading at suitable times in different ecological zones. However, gene combinations that can maximize crop yields may not exist in natural populations. Here, we planted a series of heading date mutants that harbored different heading mutant gene combinations generated by CRISPR/Cas9 gene editing technology, along with a collection of commercial varieties, across a large latitude gradient to evaluate the major effects of heading date genes and preferable gene combinations for each area. The relationship between yield and heading date was investigated. According to the pattern obtained from gene editing mutants, we concluded that the growth period of commercial varieties could be adjusted to achieve maximum yield performance in some areas. By combining the long vegetative growth allele and weak photoperiod sensitivity allele, we pinpointed an optimal balance between growth period and yield production, resulting in new partially determinate heading date to maximum yields and improved adaptability. We propose that harnessing mutations in the florigen pathway to customize the balance between vegetative and reproductive growth offers a broad toolkit for boosting crop productivity.





Similar content being viewed by others
Data availability
The datasets supporting the conclusions of this article are included within the article and its additional files. The seeds of mutants and varieties are available from the corresponding author on reasonable request.
References
Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19(9):1655–1664. https://doi.org/10.1101/gr.094052.109
Cui Y, Zhu M, Xu Z, Xu Q (2019) Assessment of the effect of ten heading time genes on reproductive transition and yield components in rice using a CRISPR/Cas9 system. Theor Appl Genet 132(6):1887–1896
Deng N, Grassini P, Yang H, Huang J, Cassman KG, Peng S (2019) Closing yield gaps for rice self-sufficiency in China. Nat Commun 10(1):1725. https://doi.org/10.1038/s41467-019-09447-9
Doebley JF, Gaut BS, Smith BD (2006) The molecular genetics of crop domestication. Cell 127(7):1309–1321
Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A (2004) Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev 18(8):926–936
Du A, Tian W, Wei M, Yan W, He H, Zhou D, Huang X, Li S, Ouyang X (2017) The DTH8-Hd1 module mediates day-length-dependent regulation of rice flowering. Mol Plant 10(7):948–961. https://doi.org/10.1016/j.molp.2017.05.006
Endo-Higashi N, Izawa T (2011) Flowering time genes heading date 1 and early heading date 1 together control panicle development in rice. Plant Cell Physiol 52(6):1083–1094
Fang J, Zhang F, Wang H, Wang W, Zhao F, Li Z, Sun C, Chen F, Xu F, Chang S, Wu L, Bu Q, Wang P, Xie J, Huang X, Zhang Y, Zhu X, Han B, Deng X, Chu C (2019) Ef-cd locus shortens rice maturity duration without yield penalty. Proc Natl Acad Sci U S A 116(37):18717–18722. https://doi.org/10.1073/pnas.1815030116
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2(4):618–620
Hosoi N (1981) Studies on meteorological fluctuation in the growth of rice plants. V. Regional differences of thermo-sensitivity, photosensitivity, basic vegetative growth and factors determining the growth duration of Japanese varieties. Jpn J Breed 31:239–250
Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O (2013) DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31(9):827–832
Ishikawa R, Aoki M, Kurotani K, Yokoi S, Shinomura T, Takano M, Shimamoto K (2011) Phytochrome B regulates heading date 1 (Hd1)-mediated expression of rice florigen Hd3a and critical day length in rice. Mol Genet Genomics 285(6):461–470. https://doi.org/10.1007/s00438-011-0621-4
Izawa T, Oikawa T, Tokutomi S, Okuno K, Shimamoto K (2000) Phytochromes confer the photoperiodic control of flowering in rice (a short-day plant). Plant J 22(5):391–399. https://doi.org/10.1046/j.1365-313x.2000.00753.x
Khush GS (1997) Origin, dispersal, cultivation and variation of rice. Plant Molecular Biology 35:25–34
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25(15):1966–1967
Li W, Zhu Z, Chern M, Yin J, Yang C, Ran L, Cheng M, He M, Wang K, Wang J (2017) A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell 170(1):114–126
Li X, Wu L, Wang J, Sun J, Xia X, Geng X, Wang X, Xu Z, Xu Q (2018) Genome sequencing of rice subspecies and genetic analysis of recombinant lines reveals regional yield- and quality-associated loci. BMC Biol 16(1):102. https://doi.org/10.1186/s12915-018-0572-x
Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8(8):1274–1284
Monfreda C, Ramankutty N, Foley JA (2008) Farming the planet: 2. Geographic distribution of crop areas, yields, physiological types, and net primary production in the year 2000. Glob Biogeochem Cycles 22(1):GB1022
Nishimura A, Aichi I, Matsuoka M (2006) A protocol for Agrobacterium-mediated transformation in rice. Nature Protocols 1(6):2796–2802
Park SJ, Jiang K, Tal L, Yichie Y, Gar O, Zamir D, Eshed Y, Lippman ZB (2014) Optimization of crop productivity in tomato using induced mutations in the florigen pathway. Nat Genet 46(12):1337–1342
Saito H, Okumoto Y, Yoshitake Y, Inoue H, Yuan Q, Teraishi M, Tsukiyama T, Nishida H, Tanisaka T (2011) Complete loss of photoperiodic response in the rice mutant line X61 is caused by deficiency of phytochrome chromophore biosynthesis gene. Theor Appl Genet 122(1):109–118
Saito H, Ogiso-Tanaka E, Okumoto Y, Yoshitake Y, Izumi H, Yokoo T, Matsubara K, Hori K, Yano M, Inoue H, Tanisaka T (2012) Ef7 encodes an ELF3-like protein and promotes rice flowering by negatively regulating the floral repressor gene Ghd7 under both short- and long-day conditions. Plant Cell Physiol 53(4):717–728. https://doi.org/10.1093/pcp/pcs029
Sudhir K, Glen S, Koichiro T (2016) MEGA7: Molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Takahashi Y, Teshima KM, Yokoi S, Innan H, Shimamoto K (2009) Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice. Proc Natl Acad Sci 106(11):4555–4560
Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K (2007) Hd3a protein is a mobile flowering signal in rice. Science 316(5827):1033–1036
Tanisaka T, Inoue H, Uozu S, Yamagata H (1992) Basic vegetative growth and photoperiod sensitivity of heading-time mutants induced in rice. Jpn J Breed 42:657–668
Wei X, Xu J, Guo H, Jiang L, Chen S, Yu C, Zhou Z, Hu P, Zhai H, Wan J (2010) DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol 153(4):1747–1758
Xu Q, Saito H, Hirose I, Katsura K, Yoshitake Y, Yokoo T, Tsukiyama T, Teraishi M, Tanisaka T, Okumoto Y (2014) The effects of the photoperiod-insensitive alleles, se13, hd1 and ghd7, on yield components in rice. Mol Breed 33(4):813–819
Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X, Zhang Q (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40(6):761–767
Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, Sasaki T (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12(12):2473–2484
Ye J, Niu X, Yang Y, Wang S, Xu Q, Yuan X, Yu H, Wang Y, Wang S, Feng Y (2018) DivergentHd1,Ghd7, and DTH7 alleles control heading date and yield potential of Japonica rice in Northeast China. Front Plant Sci 9:35
Yokoo T, Saito H, Yoshitake Y, Xu Q, Asami T, Tsukiyama T, Teraishi M, Okumoto Y, Tanisaka T (2014) Se14, encoding a JmjC domain-containing protein, plays key roles in long-day suppression of rice flowering through the demethylation of H3K4me3 of RFT1. PLoS One 9(4):e96064
Yoshitake Y, Yokoo T, Saito H, Tsukiyama T, Quan X, Zikihara K, Katsura H, Tokutomi S, Aboshi T, Mori N, Inoue H, Nishida H, Kohchi T, Teraishi M, Okumoto Y, Tanisaka T. (2015) The effects of phytochrome-mediated light signals on the developmental acquisition of photoperiod sensitivity in rice. Sci Rep 5:7709
Yuan Q, Saito H, Okumoto Y, Inoue H, Nishida H, Tsukiyama T, Teraishi M, Tanisaka T (2009) Identification of a novel gene ef7 conferring an extremely long basic vegetative growth phase in rice. Theor Appl Genet 119(4):675–684
Zhang J, Zhou X, Yan W, Zhang Z, Lu L, Han Z, Zhao H, Liu H, Song P, Hu Y (2015) Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice. New Phytol 208(4):1056–1066
Zhang Z, Hu W, Shen G, Liu H, Hu Y, Zhou X, Liu T, Xing Y (2017) Alternative functions of Hd1 in repressing or promoting heading are determined by Ghd7 status under long-day conditions. Sci Rep 7(1):5388. https://doi.org/10.1038/s41598-017-05873-1
Zhang B, Liu H, Qi F, Zhang Z, Xing Y (2019a) Genetic interactions among Ghd7, Ghd8, OsPRR37 and Hd1 contribute to large variation in heading date in rice. Rice 12:48
Zhang Z, Zhang B, Qi F, Wu H, Xing Y (2019b) Hd1 function conversion in regulating heading is dependent on gene combinations of Ghd7, Ghd8, and Ghd7.1 under long-day conditions in rice. Mol Breed 39(7):92
Zhao J, Chen H, Ren D, Tang H, Qiu R, Feng J, Long Y, Niu B, Chen D, Zhong T, Liu YG, Guo J (2015) Genetic interactions between diverged alleles of early heading date 1 (Ehd1) and heading date 3a (Hd3a)/rice flowering locus T1 (RFT1) control differential heading and contribute to regional adaptation in rice (Oryza sativa). New Phytol 208(3):936–948. https://doi.org/10.1111/nph.13503
Acknowledgments
The authors would like to thank the Excellent Youth Science Foundation of Natural Science Foundation of Liaoning Province (2019-YQ-06), Innovative talents support program of Liaoning province (LCR2018026), and Shenyang youth science and technology project (RC200452) for supporting this study.
Funding
The Excellent Youth Science Foundation of Natural Science Foundation of Liaoning Province (2019-YQ-06), Innovative talents support program of Liaoning province (LCR2018026), and Shenyang youth science and technology project (RC200452) supported this study.
Author information
Authors and Affiliations
Contributions
QX and ZX conceived and designed the research. YC organized and performed most of the experiments. QX analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(PPTX 1664 kb)
Rights and permissions
About this article
Cite this article
Cui, Y., Xu, Z. & Xu, Q. Elucidation of the relationship between yield and heading date using CRISPR/Cas9 system-induced mutation in the flowering pathway across a large latitudinal gradient. Mol Breeding 41, 23 (2021). https://doi.org/10.1007/s11032-021-01213-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-021-01213-4