Abstract
Polyploids generally show vegetative growth superiority, higher biological yield, and enhanced resistance compared with diploids, which is already an important strategy in plant breeding; however, the mechanism by which these polyploid characteristics are formed remains unclear. Here, we developed a poplar polyploidy system to compare the phenotype and gene expression differences among newly synthesized Populus allotriploids and allotetraploids and their full-sib diploids. Analysis of gene expression revealed 156 genes that were significantly differentially expressed between diploids and triploids, among which 87 and 69 were up- and downregulated, respectively. Meanwhile, 384 genes were differentially expressed between diploids and tetraploids, and there were 259 upregulated and 125 downregulated genes, respectively. Polyploids showed a bigger stomatal, and stomatal length and width increased with ploidy level, whereas relative stomatal density decreased, which may mainly attribute to the differently expression of mitogen-activated protein kinase gene, YODA, and five plant hormone-related genes (SAUR, MES17, IAR3, BAS1, ABCG25). Plant height, leaf area, and petiole length were also observed to be significantly superior in polyploids than diploids, and E3 ligase BIG BROTHER (BB) and growth-regulating factor (GRF10) were selected as candidate genes responsible for variations in organ size and leaf growth. These results provided a robust resource for investigating the function of genes associated with polyploid genotype differences. Meanwhile, our study suggests that the poplar polyploidy system with clear and complete genetic backgrounds may be used as a good model for studies in polyploidization advantage.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Polyploidization has long been recognized as a prominent force in the evolution of plant species and is an important contributor to speciation (Wood et al. 2009). It is estimated that about 70% of angiosperms have undergone at least one chromosome-doubling event during evolution (Masterson 1994). Recent developments in comparative genomics and whole genome sequencing technologies have facilitated further characterization of several previously considered diploid plant species as ancestral polyploids that have undergone diploidization by genetic recombination, such as rice (Tian et al. 2005) and the model plant Arabidopsis thaliana (Blanc et al. 2000). In addition, several diploid flowering plants are now classified as ancient polyploids (Blanc and Wolfe 2004). This evidence strongly suggests that polyploidization is a common mechanism in plant evolution. Due to the doubled chromosome number, polyploids have greater growth advantages than diploids, as characterized by larger morphological features, more metabolites, and an enhanced capacity to adapt (Chen 2010).
Allopolyploids, which have been induced by hybridization with unreduced gametes, can effectively combine polyploid advantage and heterosis, which refers to the phenomenon that hybrid offspring show greater biomass, faster growth, and better fertility than both parents (Birchler et al. 2010). Genome stability in a novel synthetic allopolyploid is thus a major challenge to survival because of the interaction between heterozygosity and ploidy effect (genomic shock). To achieve stability, a newly formed polyploid undergoes extensive genetic and epigenetic changes, which include gene reorganization, sequence loss, DNA methylation and gene silencing, and dormant transposon activation.
Populus is the model for xylophytes, and its genome is sequenced because it is economically and ecologically valuable, such as fast growing, undergoes prolific sexual reproduction, easily propagates, a small genome, and exhibits strong correlations between physiological characteristics and biomass (Tuskan et al. 2006). Furthermore, because of its features of fixed heterosis and polyploidy, triploid poplars have numerous advantages than their corresponding diploids, including greater growth rate, larger leaf area, better quality of timber, higher stress resistance, and lower fertility, which can be utilized in investigations in economic development and ecological improvement (Liao et al. 2016). As an indigenous tree, P. simonii is widely distributed in the northern areas of China. Because of its strong adaptability and relatively high resistance, it has been employed as an important pioneer afforestation species in the Three-North Shelter Forest Program, which is the largest ecological program in the world. Section Aigeiros is widely distributed around the world and an important commercial tree species for timber and biofuel; it exhibits additional advantages in reproduction and growth (Xi et al. 2012). Therefore, efforts in crossbreeding P. simonii and members of the poplar section Aigeiros may generate novel forest tree varieties, showing rapid growth and excellent timber quality as well as high levels of adaptability and resistance.
Studies involving polyploid breeding in poplar have employed stomatal size, stomatal density, and the number of chloroplast in guard cells as the most appropriate indicators in preliminary ploidy screening in sections Tacamahaca, Aigeiros, and Leuce Duby (Guo et al. 2017a, b; Lu et al. 2013; Wang et al. 2013; Xi et al. 2012), particularly for the screening of a large number of seedlings. Previous research has shown that with increasing chromosome ploidy, the size of stomata and the number of chloroplasts are enhanced, which is accompanied by a decrease in stomatal density; however, details on the genetic mechanism underlying these features are limited, mainly due to a lack of prime materials.
In the present study, we used transcriptome sequencing to investigate gene expression changes caused by hybridization and polyploidization in newly synthesized Populus diploid F1, allotriploids driven by 2n female gametes, and allotetraploids driven by heterozygotic embryo chromosome doubling. All poplars of different ploidy levels were the full-sib offspring and have exact progenitors. In this work, we seek to test the hypothesis that allotriploid and allotetraploid poplars, with the same diploid parents but showing differences in terms of stomatal size and density, leaf area, and plant height, exhibit significant changes in gene expression compared with that of full-sib diploids. In addition, our hypothesis predicts that some differentially expressed genes (DEGs) in Populus allotriploid or allotetraploid seedlings may be involved in important biological processes that are associated with stomatal development, growth, and development, thereby leading to enhanced biological processes.
Materials and methods
Plant material
Synthetic triploids and tetraploids of P. simonii (as the maternal parent, 2n = 2× = 38) and P. simonii × (P. pyramidalis + S. matsudana) (as the paternal parent, 2n = 2× = 38) as well as the corresponding diploid F1 hybrids were generated as previously described (Guo et al. 2017a, b). Briefly, triploids were induced by hybridization with post-meiotic restitution (PMR)-type 2n female gametes, and tetraploids were induced by heterozygotic embryo chromosome doubling using high temperature at 168 h after pollination. The ploidy levels of these plant materials were determined by flow cytometry.
Measurement of phenotypic traits
One-year-old cuttings were used in this study. Shoots approximately 15 cm in length were cut in March and planted in nutrient-supplemented soil (soil:vermiculite:peat:perlite = 2:1:1:1, v/v) in plastic pots. All plants were grown in the greenhouse of the Chinese Academy of Forestry (Beijing, China). After 5 months of growth, phenotypic traits, including the fourth leaf area, plant height, and petiole length of the third, fourth, sixth, seventh leaves, were measured in August, and 30 plants of each genotype were measured.
The leaves for examining stomatal characteristics were randomly selected from five plants of each genotype, respectively. The method was performed according to the method described in previous report (Guo et al. 2017a, b). The specific operation was that “The lower epidermis of the mature leaves (third or fourth leaf from the top of the plant) was removed using tweezers, placed on a glass slide with a drop of water, and flattened into the water drop with a dissecting needle. The preparations were observed under an Olympus BX43 microscope and photographed with an Olympus DP73 camera system.” At least 30 random fields were selected to count the number of stomata and to measure the length and width of stomata per field. All data were analyzed using GraphPad Prism to calculate and assess differences among different ploidy poplars.
Sampling, RNA extraction, library preparation, and RNA-Seq analysis
Fully expanded leaves (third or fourth leaf from the top of the plant) were collected separately in the morning in August. The leaves to be used for transcriptional sequencing were immediately frozen in liquid nitrogen and stored at − 80 °C until analysis. Total RNA was extracted separately from three biological replicate individuals of diploids, triploids, and tetraploids. Total RNA was extracted from each sample using TRIzol® (Invitrogen, Carlsbad, CA, USA). A total amount of 1-μg RNA from each sample was used as input material for Illumina high-throughput sequencing (RNA-Seq). Sequencing libraries were generated using NEBNext UltraTM RNA Library Prep Kit for Illumina (NEB, USA) following the manufacturer’s recommendations, and index codes were added to attribute sequences to each sample. Library quality was assessed on the Agilent Bioanalyzer 2100 system. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v4-cBot-HS according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina Hiseq 2500 platform, and paired-end reads were generated.
Clean reads were obtained by removing reads containing adapter, reads containing ploy-N, and low-quality reads from raw data. At the same time, Q20 (percentage of sequences with sequencing error rates < 1%), Q30 (percentage of sequences with sequencing error rates < 0.1%), GC-content, and sequence duplication level of the clean data were calculated. All the downstream analyses were based on clean data with high quality. The adaptor sequences and low-quality sequence reads were removed from the datasets. Raw sequences were transformed into clean reads after data processing. These clean reads were then mapped to the reference genome sequence. Only reads with a perfect match or one mismatch were further analyzed and annotated based on the reference genome. TopHat2 tools software was used to map with reference genome.
Gene function was annotated based on the following databases: Nr (NCBI non-redundant protein sequences); KOG (euKaryotic Ortholog Groups); COG (Clusters of Orthologous Groups of proteins); Swiss-Prot (a manually annotated and reviewed protein sequence database); KEGG (KEGG Ortholog database); and GO (Gene Ontology).
Differential expression analysis
Gene expression levels were estimated by fragments per kilobase of transcript per million fragments mapped (FPKM). Differential expression analysis of two conditions/groups was performed using the DESeq R package (1.10.1). DESeq provides statistical routines for determining differential expression in digital gene expression data using a model based on a negative binomial distribution. The resulting P values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted P value < 0.01 found by DESeq were classified as differentially expressed. A P value (FDR) < 0.01 and |log2(fold change)| ≥ 1 were set as threshold for significant differential expression. Hierarchical clustering of FPKM normalized gene expression was performed and visualized on heatmaps using HemI package (Deng et al. 2014) or Origin.
To validate transcript abundance as reflected by FPKM values for the assembled transcripts, 10 randomly selected genes were subjected to semi-quantitative RT-qPCR analysis to estimate relative expression levels, with the 18S gene as internal control. Three independent triplicates were performed to test the reproducibility of the amplifications. The primer sequences are listed in Table S1.
Functional annotation and pathway analysis
GO enrichment analysis of the DEGs was implemented using the GOseq R packages based Wallenius non-central hyper-geometric distribution (Young et al. 2010), which can adjust for gene length bias in DEGs. Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa et al. 2008) is a database resource for understanding high-level functions and utilities of the biological system, such as the cell, the organism, and the ecosystem from molecular-level information, particularly large-scale molecular datasets generated by genome sequencing and other high-throughput experimental technologies. We used the KOBAS (Mao et al. 2005) software to test the statistical enrichment of DEGs in the KEGG pathways.
Results
Phenotypic differences among diploids, triploids, and tetraploids
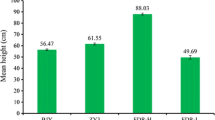
We performed phenotypic observation of three different ploidies of full-sibling hybrid progenies, including plant height, petiole length, leaf length, leaf width, stomatal length, stomatal width, and stomatal number in the unit area (Fig. 1, Table S2). The plant height of 30 plants of each genotype was measured. Diploids exhibited the lowest average height (45 cm), triploids showed the highest average height (62 cm), and tetraploids were of intermediate height (53.67 cm) (Fig. 1a, Table S2). Plant height was significantly different (P < 0.05) between diploids and triploids, whereas no significant differences were observed between diploids and tetraploids as well as between triploids and tetraploids.
Phenotypic analysis of different poplar ploidies (a bars = 5 cm; b bars = 1 cm; c bars = 20 μm). a Diploid, triploid, tetraploid plants (left to right). b Leaf from the fourth internode of diploid, triploid, tetraploid plants (left to right). c Stomatal analysis of diploid, triploid, tetraploid plants (left to right). d Phenotypic analysis of hybrid diploids, triploids, and tetraploids. The yellow arrow in c shows different stomatal guard cells among various ploidy poplars, and the red arrow indicates the enlargement of a single stomata. Different letters above the bars in d indicate statistically significant differences (P < 0.05). Abbreviations: PH, plant height; PL, petiole length; LA, leaf area; SD, stomatal density; SL, stomatal length; SW, stomatal width; SLWR, stomatal length-width ratio; SLMW, stomatal length multiplied by width
Leaf type and petiole length of diploids, triploids, and tetraploids were intermediate to those of the parents. The leaf blades of maternal parent P. simonii were nearly obovate, and petiole length was very short (Fig. S1a). The leaf blades of paternal parent P. simonii × (P. pyramidalis + S. matsudana) were nearly orbicular, but the apex acuminate, and petiole length was about 2 cm (Fig. S1b). The leaf blades of diploids, triploids, and tetraploids were closer to elliptical, and petiole length of the hybrids was also intermediate to or greater than that of the parents (Fig. 1b). The area of the fourth leaves of diploids, triploids, and tetraploids were 21.27, 44.00, and 36.65 cm2, respectively (Table S2). The leaf area of triploids and tetraploids was significantly different (P < 0.05) from that of the diploids, although there was no statistically significant between triploids and tetraploids. The average petiole length of the diploids was 1.50 cm. Triploids exhibited the longest average petiole length (4.73 cm), followed by tetraploids, which showed an average petiole length of 3.47 cm (P < 0.05).
Stomatal length, stomatal width, and stomatal number per unit area were measured and used in the calculation for relative leaf stomatal density (Fig. 1c). Stomatal length and width increased with ploidy level, whereas relative stomatal density decreased. For example, the stomatal length of diploids, triploids, and tetraploids was 23.61, 30.80, and 32.76 μm, respectively, whereas stomatal density was 22.30, 14.50, and 9.606 (Table S2). Significant difference in stomatal length, width, and density was observed among diploids, triploids, and tetraploids (P < 0.05). No significant differences in stomatal length-width ratio were observed among diploids, triploids, and tetraploids, suggesting similarities in stomatal shape. So, the stomatal length multiplied by width was roughly equal to the area of the stomata, which also significantly varied during pairwise comparisons between different ploidies.
Illumina sequencing and functional annotation of all unigenes
The average number of clean reads (paired-end reads) of diploids, triploids, and tetraploids was 27,295,967, 32,239,659, and 27,334,395, respectively, representing total 78.18 Gb of clean data. The GC content of diploids, triploids, and tetraploids was 43.66, 43.67, and 43.78%, respectively. The total number of clean reads of all ploidies showed a Q30 percentage of > 92.70% (Table 1). The total single-end reads were then mapped to the reference genome of Populus trichocarpa, and 61.55, 62.94, and 61.88% of the reads in the respective diploids, triploids, and tetraploids libraries perfectly matched the reference genome, in which 86.85, 86.03, and 85.47% of these were respectively matched uniquely mapped reads (Table 1).
All unigenes were annotated based on the results of BLASTX searches against six public databases, including Nr, Swiss-Prot, KEGG, COG, GO, and KOG in poplars of different ploidies. A total of 40,247 unigenes were annotated, wherein 40,241 unigenes (99.99%) could be matched to the Nr database, 30,376 (75.47%) in the GO database, and 28,806 (71.57%) in the Swiss-Prot database, whereas only 20,198, 14,450 and 12,039 unigenes were annotated using the KOG, COG, and KEGG databases, accounting for 50.19, 35.90, and 29.91% of the total number of unigenes (Table 2).
DEG analysis
DEGs in diploids vs. triploids, diploids vs. tetraploids, and triploids vs. tetraploids were assessed. All DEGs were subjected to BLAST analysis using the Nr, Swiss-Prot, KEGG, KOG, GO, and COG databases. The results revealed that all DEGs could be annotated to the Nr database (Table S3). Figure 2a shows that a total of 156 genes were significantly differentially expressed between diploids and triploids, and only one gene was differentially expressed in all three groups comparisons (P value < 0.01 and |log2(fold change)| ≥ 1). In particular, 66 and 64 DEGs were shared between diploids and tetraploids, and triploids and tetraploids, respectively, and 27 DEGs were not shared with the other two group comparisons. A total of 384 genes were differentially expressed between diploids and tetraploids. In addition to the 66 shared DEGs between diploids and triploids, 132 DEGs were shared between triploids and tetraploids, whereas 187 DEGs were not. Furthermore, 273 genes showed significant differential expression between triploids and tetraploids, and 78 DEGs were not shared with the other two group comparisons (Fig. 2a). In all three group comparisons, the number of upregulated DEGs was higher than the number of downregulated genes. Compared with diploids, triploids showed 87 upregulated and 69 downregulated genes, whereas tetraploids had 259 upregulated and 125 downregulated genes. Furthermore, 155 genes were upregulated, and 118 genes were downregulated between triploids and tetraploids (Fig. 2b). The observed changes in gene expression may be caused by differences in ploidy levels. More DEGs were identified, and larger differences in expression were observed between diploids and tetraploids compared with that between diploids and triploids or triploids and tetraploids, which may be associated with larger differences in ploidy level.
Venn diagram and histogram of the number of DEGs in diploids, triploids, and tetraploids. “a” is the control group, and “b” is the experimental group in “a vs. b.” P value (FDR) < 0.01 and |log2 (fold change)| ≥ 1. a Venn diagram of the number of DEGs. b Histogram showing the number of DEGs that were up- or downregulated between different libraries. c Semi-quantitative RT-PCR validation of differentially expressed known gene sequences obtained from deep sequencing. Di., Tri., and Tetra. represent diploids, triploids, and tetraploids, respectively. The ordinates in the charts represent the FPKM
To validate the results of RNA-Seq analysis, we designed primers of 10 randomly selected genes for RT-PCR analysis to confirm the expression among different ploidy poplars (Fig. 2c). All genes had been successfully amplified, and their expression patterns were consistent with the results of high-throughput sequencing, indicating that our sequencing data were reliable and could be subjected to further analysis.
Functional annotation and pathway analysis of DEGs
All of the DEGs from the three group comparisons were functionally annotated using the GO database. Approximately 112 DEGs in diploids vs. triploids, 303 DEGs in diploids vs. tetraploids, and 220 DEGs in triploids vs. tetraploids were annotated using the GO database and classified into three major GO categories, namely, biological process, cellular component, and molecular function, which were then further classified into 52 functional subcategories (Fig. S2 and Table S4). In the biological process category, “cellular process,” “metabolic process,” “single-organism process,” and “response to stimulus” were the most highly represented terms. In the cellular component category, “cell part,” “cell,” and “organelle” were the prominently represented terms. In the molecular function category, “binding” and “catalytic activity” were the predominant groups. These main GO terms provided a general overview of ontology content and showed that a wide range of biological activities occur in different ploidy poplars. Some DEGs that were only observed between diploids and tetraploids or triploids and tetraploids were classified into five sub-categories, namely, “extracellular region part” (3 and 3 DEGs), “protein binding transcription factor activity” (1 and 1 DEG), “enzyme regulator activity” (6 and 4 DEGs), “nutrient reservoir activity” (2 and 3 DEGs), and “rhythmic process” (3 and 3 DEGs), respectively (Table S4).
To further categorize of these DEGs, hierarchical clustering analysis was performed for DEGs with significant trends and visualized using a heatmap (Fig. S3). Functional enrichment tests revealed that the patterns of gene expression in cluster 1 increased with higher ploidy levels, and related genes were most enriched with GO terms, such as defense response, signal transduction, response to other organism, cellular response to inorganic substance, cellular response to gibberellin stimulus, and ADP binding. The DEGs in clusters 2 and 3 showed the lowest expression levels in triploids and the highest expression levels in tetraploids and were mostly enriched with GO terms, such as post-embryonic organ development, defense response, and epidermis development. DEGs with the highest expression in triploids were annotated to GO terms, such as macromolecule biosynthetic process and cellular carbohydrate metabolic process (cluster 4), which are probably related to greater growth vigor. DEGs with the lowest expression in tetraploids were annotated to GO terms, such as positive regulation of flavonoid biosynthetic process, whereas DEGs showing the highest expression in diploids were classified under GO terms, such as oxidoreductase activity and aspartate-ammonia ligase activity.
Approximately 77 DEGs in diploids vs. triploids, 173 DEGs in diploids vs. tetraploids, and 128 DEGs in triploids vs. tetraploids were annotated by COG, which were clustered into 25 different functional categories (Fig. S4 and Table S5). COG functional annotation of DEGs of the three pairwise group comparisons indicated a similar pattern of distribution. For these categories, the highest number of DEGs was clustered into the “general functional prediction only” category, accounting for 19.64, 21.71, and 20.09% of the total number of DEGs, respectively. Among COG classes of diploids vs. triploids, the cluster for “function unknown” (16 DEGs, 14.29%) represented the second largest group, followed by “replication, recombination, and repair” (10 DEGs, 8.93%) and “transcription” (10 DEGs, 8.93%). However, in diploids vs. tetraploids, the second most enriched category was “signal transduction mechanisms” (40 DEGs, 13.16%), followed by “replication, recombination, and repair” (36 DEGs, 11.84%) and “transcription” (35 DEGs, 11.51%). Furthermore, there was a small difference in triploids vs. tetraploids in that the second largest group was “replication, recombination, and repair” (27 DEGs, 12.62%), followed by “signal transduction mechanisms” (26 DEGs, 12.15%), “function unknown” (26 DEGs, 12.15%), and “transcription” (25 DEGs, 11.68%).
KEGG pathway analysis was performed to identify the pathways in which DEGs were involved and enriched. Approximately 41 DEGs in diploids vs. triploids, 94 DEGs in diploids vs. tetraploids, and 95 DEGs in triploids vs. tetraploids were all mainly classified into five major functional categories (Table S6; Fig. 3a). Figure 3 shows that the DEGs of the three pairwise group comparisons were significantly enriched in the category of metabolism (69.00, 69.00, and 72.00%), and then the DEGs were further assigned to two major KEGG pathways, namely, amino acid metabolism (20, 11, and 13%) and carbohydrate metabolism (20, 14, and 13%), respectively (Fig. 3a). Second, the next significant category was genetic information processing (22%, 11%) in diploids vs. triploids and triploids vs. tetraploids, which included the following KEGG pathways: protein processing in the endoplasmic reticulum (ko04141), homologous recombination (ko03440), DNA replication (ko03030), nucleotide excision repair (ko03420), mismatch repair (ko03430), basal transcription factors (ko03022), RNA transport (ko03013), and ribosome (ko03010), which may be related to the structural changes in the genome that was caused by an increase in ploidy level. Meanwhile, in diploids vs. tetraploids, organismal systems (14%) were the second most significant category, which included two KEGG pathways, namely, circadian rhythm-plant (ko04712) and plant-pathogen interaction (ko04626), which belong to the environmental adaptation pathway and may be related to structural changes in the genome caused by an increase in ploidy level.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation of DEGs among diploids, triploids, and tetraploids. a Distribution of DEGs into KEGG biological categories and classification into the KEGG “metabolism” category. b Global metabolic circuit map of the DEGs. The red lines depict pathways enriched with upregulated genes. The green lines represent pathways involving downregulated genes. Significance is defined as P value (FDR) < 0.01 and |log2(fold change)| ≥ 1
Based on the KEGG pathway and the expression and functional annotation of DEGs, an overview of metabolic network pathways (Fig. 3b) among diploids, triploids, and tetraploids was further analyzed using iPath2.0 (pathways.embl.de/iPath2.cgi) (Yamada et al. 2011). The metabolic network was apparently similar in topology in both diploids vs. triploids and diploids vs. tetraploids, but not in triploids vs. tetraploids, which suggest that metabolic pathways enriched with upregulated genes in triploids and tetraploids were more than enriched with downregulated genes compared to diploids.
DEGs involved in stomatal development
Plant stomata are specialized epidermal cell structures, which formed by two guard cells surrounding a pore, serving as channels that modulate gas (CO2 and O2) and water vapor exchange between plants and their environment to maintain photosynthetic and water-use efficiency (Blatt and Grabov 1997). Stomatal distribution follows the one-cell spacing rule (Fig. 4a). Here, four genes associated with stomatal development were differentially expressed between tetraploids and diploids, with three genes being upregulated and one gene downregulated (Fig. 4). The expression of stomatal development-related genes, such as YDA, IAR3, bHLH35, was upregulated in tetraploids. Conversely, BAS1 were downregulated in tetraploids. Compared with diploids, four genes were upregulated, and one gene was downregulated in triploids (Fig. 4). Among these, stomatal development-related genes, such as methylesterase 17 (MES17), IAR3, ABCG25, bHLH35, were upregulated expression in triploids. Conversely, SAUR were downregulated in triploids. Overall, more stomatal development-related genes were upregulated in polyploids than in diploids.
Stomatal development is regulated by genetic signals and environmental factors. a Genetic control and hormonal signaling is involved in stomatal development from mother of meristemoid cells (MMC) to guard cells (GC). Green arrows indicate positive regulation, and red blunt lines indicate negative regulation. Experimentally confirmed genetic interactions are represented by solid lines, and speculated genetic interactions are indicated by dashed lines. The genes in the purple box affect stomatal development in tetraploids, the genes in the yellow box affect stomatal development in triploids, and the genes in the yellow-purple half box affect stomatal development both in triploids and tetraploids. Stages of stomatal development from MMC to GC are shown at the bottom of the figure. b Hierarchical clustering of DEGs in stomatal development. Values in the color key refer to FPKM
DEGs involved in lignin biosynthesis
Lignin biosynthesis is completed by the effect of a series of enzymes, which requires the participation of synergistic enzymes. In the present study, KEGG analyses of DEGs revealed the presence of 6 genes (CAD, CSE, 4CL, CCoAoMT, CAD, and peroxidase) involved in the biosynthesis of lignin (Fig. 5). CAD was downregulated in triploids compared to diploids, and the other five genes were equal to or higher than in diploids, especially peroxidase, which showed significantly higher expression (log2FC = 5.28) (Fig. 5). In tetraploids, CSE, 4CL, and COMT were downregulated compared with diploids, whereas other genes, such as CAD, CCoAoMT, and peroxidase, were highly expressed (Fig. 5).
Lignin biosynthesis of phenylpropanoid biosynthesis pathway study by KEGG analysis showing the different identified enzymes. Value of the color key refers to the FPKM. The relative expression of each gene in the figure is represented by three squares that correspond to diploids, triploids, and tetraploids, respectively
Discussions
Different Populus polyploid populations are a good model for studies in polyploidization and heterosis advantage
The materials used in this study have clear and complete genetic backgrounds, including the male parent, female parent, diploid full-sibs, allotriploids, and allotetraploids. The inheritance and variations of gene expression caused by hybridization and allopolyploidization can be identified, by comparing the gene expression patterns among diploid F1, allotriploid, and allotetraploid, and their parents, which suggest that these different ploidy poplars are a good model for studies in polyploidization and heterosis advantage, genomic variations, and stability after polyploidization.
In the present study, differences in gene expression patterns were observed in different ploidy poplars, which suggests that chromosome doubling has imparted genome shock to novel synthesized poplar polyploids and affected the expression of some genes to a certain extent, thereby affecting the phenotypic properties of poplar polyploids, such as growth traits, environmental resilience, and resistance. The largest gene expression difference was observed between diploid and tetraploids, which may be associated with relatively larger ploidy differences, followed by differences in gene expression between triploids and tetraploids; the gene expression differences between diploids and triploids were the smallest and may be related to the mechanism underlying the formation of triploids. There are three modes of forming 2n female gametes, including first division restitution (FDR), second division restitution (SDR), and PMR, which are derived from embryo sac chromosome doubling. Previous study (Cheng et al. 2015) has observed differences in gene expression between the allopolyploids driven by three types of 2n female gametes and the diploid F1 and determined that the number of DEGs between the triploid-FDR and diploid F1 (1140 DEGs) was far more than that between the triploid-PMR and diploid F1 (55 DEGs) and between triploid-SDR and diploid F1 (32 DEGs). In our investigation, all triploid poplars originated from the PMR type 2n female gametes, which are possibly characterized by complete homozygosity, and this may result in minimal gene expression differences between diploids and triploids. Nevertheless, most of the DEGs between diploids and PMR-type triploids were annotated to metabolic pathways, such as carbohydrate and lipid metabolism, which also reflects that triploids have significant advantages in carbon metabolism and utilization efficiency over diploids, which in turn may be responsible for the fast-growing trait observed in triploids.
Newly formed allopolyploids and autopolyploids exhibit considerable meiotic complexity, such as multivalent pairing and unbalanced gametes, resulting in reduced fertility. Previous studies found that polyploidization strengthens F1 pollen sterility loci interactions, resulting in meiosis abnormalities and pollen sterility in autotetraploid rice, and specifically expressed miRNAs, especially meiosis-related miRNAs, and TEs-siRNAs were detected in autotetraploid rice (Li et al. 2016, 2017; Wu et al. 2015). Guo et al. (2017a, b) successfully cultivated neo-tetraploid rice with more than 80% seed setting in F5 by expanding the distance of parental relationship and continuously self-crossed and revealed some genes that may associate with the fertility and heterosis in neo-tetraploid rice. The above results provide a favorable and scientific reference for us to study the improvement of allopolyploid fertility.
Pattern of stomatal development in polyploids
Stomatal development patterns, particularly stomatal density, are regulated by genetic signals (Bergmann 2004; Sugano et al. 2010) and hormones, such as auxin, BR, and ABA (Kim et al. 2012; Tanaka et al. 2013; Zhang et al. 2014). Genetically defined signal transduction cascades that negatively regulate the basal pathway of stomatal lineage, including peptide ligands or proteinases (EPF1/2, SDD1), receptor proteins (TMM, ER family), and a MAPK module (Lampard et al. 2008; Torii 2012; Wang et al. 2007). Interestingly, another peptide ligand, STOMAGEN, and the bHLH transcription factors positively regulate stomatal development (Lampard et al. 2008; Sugano et al. 2010).
Despite extensive efforts to elucidate the regulatory mechanism of stomatal development, our understanding of the mechanism by which genetic signals and environmental factors modulate polyploidy stomatal development patterns remains unclear. In the present study, significant changes in the expression of some genes related to stomatal development were observed in polyploids compared with diploids. Based on previous findings and the observed variations in the expression of the genes associated with stomatal development in this study, we propose a mechanism for the formation of polyploid stomatal patterns (Fig. 4a).
In tetraploids, upregulated expression of YODA (YDA) was observed compared to diploids (log2FC = 1.74) (Fig. 4b). In stomatal development signaling pathways, the MAPKKK YDA plays a central role in controlling cell identities in the epidermis by acting as a cell-fate switch and negatively regulating stomatal development; loss-of-function mutations lead to an excessive number of cells adopting a stomatal fate and elevate stomatal density (Bergmann et al. 2004; Wang et al. 2007). The specific and high expression of YDA in tetraploids indicates that YDA plays a major role in the occurrence of lower stomatal density in tetraploids. Previous study has indicated that auxin negatively regulates stomatal development through binding the STOMAGEN promoter to suppress its expression and then inhibit stomatal development (Zhang et al. 2014). IAR3 encodes IAA-amino acid hydrolases that can cleave IAA-amino acid conjugates to release free IAA, and triple hydrolase mutant seedlings have lower IAA levels than the wild-type (Rampey et al. 2004). In our study, IAR3 is expressed at a higher level than diploids (Fig. 4b), potentially resulting in enhanced endogenous levels of auxin that inhibit stomatal development. Moreover, BAS1 encodes a cytochrome P450 monooxygenase that inactivates Brassinosteroids (BRs) (Turk et al. 2005), and research indicates that BRs inhibit stomatal development by activating YDA, and fewer stomata were observed in plants overexpressing some of the positive BR-signaling components (Kim et al. 2012). In our study, the expression of BAS1 was significantly decreased in tetraploids compared to diploids (Fig. 4b), potentially resulting in enhanced endogenous levels of BRs and inhibition of stomatal development. These findings suggest that YDA and the phytohormones auxin and BRs, and their interactions play essential roles in the specific stomatal pattern of tetraploids (Fig. 4a).
In triploids, there are no significant DEGs in the genetically defined signal transduction cascade of stomatal development, which suggests that the signaling cascades of stomatal development in triploids are more unpredictable because of its unique genome structure. However, the expression of the early auxin response gene SAUR significantly decreased, and MES17 is expressed at higher levels in triploids compared with both diploids and tetraploids. IAR3 was also upregulated in triploids (Fig. 4b). Among them, SAUR modulates polar auxin transport, thereby affecting auxin content (Ren and Gray 2015), MES17 efficiently and specifically hydrolyzes MeIAA to IAA (Yang et al. 2008), and IAR3 promotes the release of free IAA. Variations in this gene expression potentially result in an enhancement of endogenous levels of auxin, which then inhibit stomatal development (Zhang et al. 2014). Moreover, previous study suggests that abscisic acid (ABA) induces stomatal closure and limits initiation of stomatal development, ABA-deficient aba2-2 mutants exhibit an increase in the proportion of stomata within a smaller cotyledon (Tanaka et al. 2013), and overexpression of ABCG25 enhances ABA signals (Kuromori et al. 2016). ABCG25 was upregulated in triploids compared to diploids and tetraploids (Fig. 4b), potentially resulting in enhanced ABA levels in guard cells and inhibition of stomatal development. These findings suggest that the phytohormones auxin and ABA play essential roles in the specific stomatal pattern of triploids (Fig. 4a).
Another noteworthy phenomenon is the expression of bHLH35, which belongs to bHLH subfamily III; it significantly increased with higher ploidy levels, suggesting that it could also play essential roles in the specific stomatal patterns of triploids and tetraploids (Fig. 4b).
DEGs associated with phenotypic traits
Final organ size was tightly regulated in species-specific, but the mechanisms are not well understood, despite their central importance. The E3 ligase BIG BROTHER (BB), with E3 ubiquitin-ligase activity that represses plant organ growth, negatively regulated the duration of cell proliferation in leaves and petals (Disch et al. 2006). BB limits organ size in a dosage-dependent manner and independently of the major phytohormones. Lack of BB activity resulted in larger organs. Conversely, overexpression expression of BB formed smaller organs. In our study, the expression of BB was upregulated both in diploids vs. triploids (log2FC = 4.22) and diploids vs. tetraploids (log2FC = 4.52), but at the same time, the leaf area and plant height of triploids and tetraploids were significantly higher than in diploids (Fig. 1). One possible explanation for this discrepancy is that BB is a species-specific size checkpoint, which allows plants to grow until they reach a final predetermined level, and then terminates organ growth according to species-specific intrinsic information about final size, suggesting that polyploids may need higher BB expression levels to attain normal organ sizes.
The growth-regulating factor (GRF) gene family plays crucial roles in the control of leaf growth. In previous study (Wu et al. 2014), the overexpression of ZmGRF10 resulted in a 6% decrease in plant height and a 10% reduction in leaf size through decreasing cell proliferation in maize. In our study, the expression of GRF10 was downregulated both in diploids vs. triploids (log2FC = − 3.72) and diploids vs. tetraploids (log2FC = − 3.16), which suggests that the lower level expression of GRF10 in triploids and tetraploids may lead to an increase in leaf size and plant height in triploids and tetraploids (Fig. 1).
Previous study has shown that transient AtGR1 accumulation mediates cell cycle arrest before mitosis (Deveaux et al. 2000), which is necessary for DNA repair prior to chromosome segregation and cytokinesis. In our study, the expression of GR1 was upregulated both in triploids and tetraploids compared with diploids, which may be associated with their larger genomes and greater amounts of DNA repair in polyploids.
CUT1, which encodes a VLCFA-condensing enzyme, was required for elongation of C24 fatty acids, which is an essential step of cuticular wax production, and contributes to suberin biosynthesis. Sense suppression of CUT1 results in waxless stems and siliques in transgenic Arabidopsis plants (Millar et al. 1999). In our study, CUT1 expression was upregulated in tetraploids compared to both diploids (log2FC = 5.45) and triploids (log2FC = 4.71), which might imply that the wax content of tetraploid epidermis was likely to change compared with diploids and triploids.
EXO70A1 is involved in polarized cell growth and organ morphogenesis (Fendrych et al. 2010). exo70A1 mutants have a defect in cell plate assembly and have dramatically reduced fertility. In our studies, EXO70A1 expression in triploids was downregulated to 0.003 (FPKM), whereas it was 2.17 in diploids and 0.46 in tetraploids, which might lead to defects in the initial cell plate assembly phases, and normal cell division cannot be completed, resulting in reduced fertility in triploids.
Lignin, a phenolic polymer deposited in the secondary cell wall, is an important substance in the metabolic pathway of phenylpropanoids. Because of their hydrophobic and strengthening nature, deposition of lignin enables xylem vessels to transport water and nutrients, reinforces plant cell walls, allows vascular plants to grow tall and stand upright, and acts as a mechanical barrier to pathogens (Boudet 2007). In the present study, six genes involved in the biosynthesis of lignin were differentially expressed between polyploids and diploids (Fig. 5). These genes have been identified, and their roles have been investigated (Table S7). In previous study, COMT silencing or downregulated expression would lead to near total loss or reduction in S lignin (Guo et al. 2001). Meanwhile, a cse mutant deposits less lignin due to S lignin decreases (Vanholme et al. 2013). Moreover, the inhibition or downregulated expression of one of the four genes (4CL, CCoAoMT, CAD, and peroxidase) would cause a decline in lignin production (Li et al. 2003, 2009; Lu et al. 2004; Zhong et al. 2000). In our study, CAD was the only gene that showed a little lower expression in triploids compared with diploids, but there was no significant difference. The expression of the other five genes was equal to or higher than in diploids (Fig. 5), which might indicate that the lignin content in triploids was higher than that in diploids. In tetraploids, there was no significant trend in gene expression, which might not be able to predict the change trend of lignin content in tetraploids. DEG analyses of lignin biosynthesis-related genes indicate the importance of the lignin biosynthesis pathway in poplar polyploidization processes, suggesting that polyploidization can alter lignin content and components.
In conclusion, we developed and compared transcriptomic differences among three different ploidy poplars, and this work represents a genome-wide study to characterize transcriptional responses to polyploidization. This study may serve as a resource for further polyploid studies and gene functions in relation to polyploid advantage to better understand the molecular regulatory mechanisms of stomatal development in polyploidy poplars.
References
Bergmann DC (2004) Integrating signals in stomatal development. Curr Opin Plant Biol 7:26–32. https://doi.org/10.1016/j.pbi.2003.10.001
Bergmann DC, Lukowitz W, Somerville CR (2004) Stomatal development and pattern controlled by a MAPKK kinase. Science 304:1494–1497. https://doi.org/10.1126/science.1096014
Birchler J, Yao H, Chudalayandi S, Vaiman D, Veitia RA (2010) Heterosis. Plant Cell 22:2105–2112. https://doi.org/10.1105/tpc.110.076133
Blanc G, Barakat A, Guyot R, Cooke R, Delseny M (2000) Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell 12:1093–1101. https://doi.org/10.1105/tpc.12.7.1093
Blanc G, Wolfe KH (2004) Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16:1667–1678. https://doi.org/10.1105/tpc.021345
Blatt MR, Grabov A (1997) Signalling gates in abscisic acid-mediated control of guard cell ion channels. Physiol Plant 100:481–490. https://doi.org/10.1111/j.1399-3054.1997.tb03052.x
Boudet AM (2007) Evolution and current status of research in phenolic compounds. Phytochemistry 68:2722–2735. https://doi.org/10.1016/j.phytochem.2007.06.012
Chen ZJ (2010) Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci 15:57–71. https://doi.org/10.1016/j.tplants.2009.12.003
Cheng S, Yang J, Liao T, Zhu X, Suo Y, Zhang P, Wang J, Kang X (2015) Transcriptomic changes following synthesis of a Populus full-sib diploid and allotriploid population with different heterozygosities driven by three types of 2n female gamete. Plant Mol Biol 89:493–510. https://doi.org/10.1007/s11103-015-0384-0
Deng W, Wang Y, Liu Z, Cheng H, Xue Y (2014) HemI: a toolkit for illustrating heatmaps. PLoS One 9:e111988. https://doi.org/10.1371/journal.pone.0111988
Deveaux Y, Alonso B, Pierrugues O, Godon C, Kazmaier M (2000) Molecular cloning and developmental expression of AtGR1, a new growth-related Arabidopsis gene strongly induced by ionizing radiation. Radiat Res 154:355–364. https://doi.org/10.1667/0033-7587(2000)154[0355:MCADEO]2.0.CO;2
Disch S, Anastasiou E, Sharma VK, Laux T, Fletcher JC, Lenhard M (2006) The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner. Curr Biol 16:272–279. https://doi.org/10.1016/j.cub.2005.12.026
Fendrych M, Synek L, Pečenková T, Toupalová H, Cole R, Drdová E, NebesářOvá J, Šedinová M, Hála M, Fowler JE (2010) The Arabidopsis exocyst complex is involved in cytokinesis and cell plate maturation. Plant Cell 22:3053–3065. https://doi.org/10.1105/tpc.110.074351
Guo D, Chen F, Inoue K, Blount JW, Dixon RA (2001) Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa. Impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 13:73–88. https://doi.org/10.1105/tpc.13.1.73
Guo HB, Mendrikahy JN, Xie L, Deng JF, Lu ZJ, Wu JW, Li X, Shahid MQ, Liu XD (2017a) Transcriptome analysis of neo-tetraploid rice reveals specific differential gene expressions associated with fertility and heterosis. Sci Rep 7:40139. https://doi.org/10.1038/srep40139
Guo L, Xu W, Zhang Y, Zhang J, Wei Z (2017b) Inducing triploids and tetraploids with high temperatures in Populus sect. Tacamahaca. Plant Cell Rep 36:313–326. https://doi.org/10.1007/s00299-016-2081-0
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36:480–484. https://doi.org/10.1093/nar/gkm882
Kim TW, Michniewicz M, Bergmann DC, Wang ZY (2012) Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482:419–422. https://doi.org/10.1038/nature10794
Kuromori T, Fujita M, Urano K, Tanabata T, Sugimoto E, Shinozaki K (2016) Overexpression of AtABCG25 enhances the abscisic acid signal in guard cells and improves plant water use efficiency. Plant Sci 251:75–81. https://doi.org/10.1016/j.plantsci.2016.02.019
Lampard GR, Macalister CA, Bergmann DC (2008) Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322:1113–1116. https://doi.org/10.1126/science.1162263
Li X, Shahid MQ, Xia J, Lu ZJ, Fang N, Wang L, Wu JW, Chen ZX, Liu XD (2017) Analysis of small RNAs revealed differential expressions during pollen and embryo sac development in autotetraploid rice. BMC Genomics 18(1):129. https://doi.org/10.1186/s12864-017-3526-8
Li X, Shahid MO, Wu JW, Wang L, Liu XD, Lu YG (2016) Comparative small rna analysis of pollen development in autotetraploid and diploid rice. Int J Mol Sci 17(4):499. https://doi.org/10.3390/ijms17040499
Li X, Ying Y, Yao J, Chen G, Li X, Zhang Q, Wu C (2009) FLEXIBLE CULM 1 encoding a cinnamyl-alcohol dehydrogenase controls culm mechanical strength in rice. Plant Mol Biol 69:685–697. https://doi.org/10.1007/s11103-008-9448-8
Li Y, Kajita S, Kawai S, Katayama Y, Morohoshi N (2003) Down-regulation of an anionic peroxidase in transgenic aspen and its effect on lignin characteristics. J Plant Res 116:175–182. https://doi.org/10.1016/S0921-0423(01)80073-3
Liao T, Cheng S, Zhu X, Min Y, Kang X (2016) Effects of triploid status on growth, photosynthesis, and leaf area in Populus. Trees 30:1137–1147. https://doi.org/10.1007/s00468-016-1352-2
Lu H, Zhao YL, Jiang XN (2004) Stable and specific expression of 4-coumarate: coenzyme A ligase gene (4CL1) driven by the xylem-specific Pto4CL1 promoter in the transgenic tobacco. Biotechnol Lett 26(14):1147–1152. https://doi.org/10.1023/B:BILE.0000035487.91628.9e
Lu M, Zhang P, Kang X (2013) Induction of 2n female gametes in Populus adenopoda Maxim by high temperature exposure during female gametophyte development. Breed Sci 63:96–103. https://doi.org/10.1270/jsbbs.63.96
Mao X, Tao C, Olyarchuk JG, Wei L (2005) Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21:3787–3793. https://doi.org/10.1093/bioinformatics/bti430
Masterson J (1994) Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science 264:421–424. https://doi.org/10.1126/science.264.5157.421
Millar AA, Clemens S, Zachgo S, Giblin EM, Taylor DC, Kunst L (1999) CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell 11:825–838. https://doi.org/10.1105/tpc.11.5.825
Rampey RA, Leclere S, Kowalczyk M, Ljung K, Sandberg G, Bartel B (2004) A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol 135:978–988. https://doi.org/10.1104/pp.104.039677
Ren H, Gray WM (2015) SAUR proteins as effectors of hormonal and environmental signals in plant growth. Mol Plant 8:1153–1164. https://doi.org/10.1016/j.molp.2015.05.003
Sugano SS, Shimada T, Imai Y, Okawa K, Tamai A, Mori M, Haranishimura I (2010) Stomagen positively regulates stomatal density in Arabidopsis. Nature 463:241–244. https://doi.org/10.1038/nature08682
Tanaka Y, Nose T, Jikumaru Y, Kamiya Y (2013) ABA inhibits entry into stomatal-lineage development in Arabidopsis leaves. Plant J 74:448–457. https://doi.org/10.1111/tpj.12136
Tian CG, Xiong YQ, Liu TY, Sun SH, Chen LB, Chen MS (2005) Evidence for an ancient whole-genome duplication event in rice and other cereals. Acta Genet Sin 32:519–527
Torii KU (2012) Mix-and-match: ligand-receptor pairs in stomatal development and beyond. Trends Plant Sci 17:711–719. https://doi.org/10.1016/j.tplants.2012.06.013
Turk EM, Fujioka S, Seto H, Shimada Y, Takatsuto S, Yoshida S, Wang H, Torres QI, Ward JM, Murthy G (2005) BAS1 and SOB7 act redundantly to modulate Arabidopsis photomorphogenesis via unique brassinosteroid inactivation mechanisms. Plant J 42:23–34. https://doi.org/10.1111/j.1365-313X.2005.02358.x
Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A (2006) The genome of black cottonwood, Populus trichocarpa (Torr.&Gray). Science 313:1596–1604. https://doi.org/10.1126/science.1128691
Vanholme R, Cesarino I, Rataj K, Xiao Y, Sundin L, Goeminne G, Kim H, Cross J, Morreel K, Araujo P (2013) Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 341:1103–1106. https://doi.org/10.1126/science.1241602
Wang H, Ngwenyama N, Liu Y, Walker J, Zhang S (2007) Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19:63–73. https://doi.org/10.1105/tpc.106.048298
Wang J, Shi L, Song S, Tian J, Kang X (2013) Tetraploid production through zygotic chromosome doubling in Populus. Silva Fenn 47:932. https://doi.org/10.14214/sf.932
Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH (2009) The frequency of polyploid speciation in vascular plants. Proc Natl Acad Sci U S A 106:13875–13879. https://doi.org/10.1073/pnas.0811575106
Wu JW, Shahid MQ, Chen L, Chen ZX, Wang L, Liu XD, Lu YG (2015) Polyploidy enhances F1 pollen sterility loci interactions that increase meiosis abnormalities and pollen sterility in autotetraploid rice. Plant Physiol 169:2700–2717. https://doi.org/10.1104/pp.15.00791
Wu L, Zhang D, Xue M, Qian J, He Y, Wang S (2014) Overexpression of the maize GRF10, an endogenous truncated growth-regulating factor protein, leads to reduction in leaf size and plant height. J Integr Plant Biol 56:1053–1063. https://doi.org/10.1111/jipb.12220
Xi X, Li D, Xu W, Guo L, Zhang J, Li B (2012) 2n egg formation in Populus × euramericana (Dode) Guinier. Tree Genet Genomes 8:1237–1245. https://doi.org/10.1007/s11295-012-0510-y
Yamada T, Letunic I, Okuda S, Kanehisa M, Bork P (2011) iPath2.0: interactive pathway explorer. Nucleic Acids Res 39:412–415. https://doi.org/10.1093/nar/gkr313
Yang Y, Xu R, Ma CJ, Vlot AC, Klessig DF, Pichersky E (2008) Inactive methyl indole-3-acetic acid ester can be hydrolyzed and activated by several esterases belonging to the AtMES esterase family of Arabidopsis. Plant Physiol 147:1034–1045. https://doi.org/10.1104/pp.108.118224
Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11:1–12. https://doi.org/10.1186/gb-2010-11-2-r14
Zhang JY, He SB, Li L, Yang HQ (2014) Auxin inhibits stomatal development through MONOPTEROS repression of a mobile peptide gene STOMAGEN in mesophyll. Proc Natl Acad Sci U S A 111:3015–3023. https://doi.org/10.1073/pnas.1400542111
Zhong R, Rd MW, Himmelsbach DS, Nd PF, Ye ZH (2000) Essential role of caffeoyl coenzyme A O-methyltransferase in lignin biosynthesis in woody poplar plants. Plant Physiol 124:563–577. https://doi.org/10.1104/pp.124.2.563
Acknowledgments
We would like to thank LetPub (www.letpub.com) for providing linguistic assistance during the preparation of this manuscript.
Funding
This work was supported by the Fundamental Research Funds for the Central Non-profit Research Institution of Chinese Academy of Forestry (CAFYBB2018QB001).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOC 3474 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Liqin, G., Jianguo, Z., Xiaoxia, L. et al. Polyploidy-related differential gene expression between diploid and synthesized allotriploid and allotetraploid hybrids of Populus. Mol Breeding 39, 69 (2019). https://doi.org/10.1007/s11032-019-0975-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-019-0975-6