Abstract
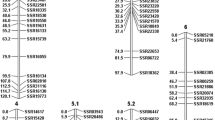
Bacterial wilt (Burkholderia caryophylli (Burkholder) Yabuuchi et al.) is one of the most damaging diseases during carnation (Dianthus caryophyllus L.) cultivation in Japan. To find molecular markers for use in marker-assisted selection, we constructed a simple sequence repeat (SSR)-based genetic linkage map of carnation using an F2 population of 90 plants derived from a cross between a highly resistant line (85-11) and a susceptible cultivar (Pretty Favvare). To develop a large number of SSR markers, we constructed four new SSR-enriched genomic libraries and conducted expressed sequence tag analysis. We mapped 178 SSR loci into 16 linkage groups. The map covered 843.6 cM, with an average distance of 6.5 cM between two loci. This is the first report of a genetic linkage map based mainly on SSR markers in the genus Dianthus. Quantitative trait locus (QTL) analysis identified one locus for resistance to bacterial wilt in linkage group (LG) B4. The locus explained 63.0% of the phenotypic variance for resistance to bacterial wilt. The SSR markers CES1161 and CES2643 that were closest to the QTL were efficient markers for selecting lines with resistance derived from line 85-11. A positional comparison using SSR markers as anchor loci revealed that LG B4 corresponded to LG A6 in a previously constructed map. We found that the position of the resistance locus derived from line 85-11 was similar to that of the major resistance locus observed for a highly resistant wild species, Dianthus capitatus ssp. andrzejowskianus.




Similar content being viewed by others
References
Abe H, Nakano M, Nakatsuka A, Nakayama M, Koshioka M, Yamagishi M (2002) Genetic analysis of floral anthocyanin pigmentation traits in Asiatic hybrid lily using molecular linkage maps. Theor Appl Genet 105:1175–1182
Asamizu E, Nakamura Y, Sato S, Fukuzawa H, Tabata S (1999) A large scale structural analysis of cDNAs in a unicellular green alga, Chlamydomonas reinhardtii. I. Generation of 3,433 non-redundant expressed sequence tags. DNA Res 6:369–373
Blair MW, Pedraza F, Buendia HF, Gaitan-Solis E, Beebe SE, Gepts P, Tohme J (2003) Development of a genome-wide anchored microsatellite map for common bean (Phaseolus vulgaris L.). Theor Appl Genet 107:1362–1374
Brownstein MJ, Carpten JD, Smith JR (1996) Modulation of non-templated nucleotide addition by Taq DNA polymerase: primer modifications that facilitate genotyping. BioTechniques 20:1004–1010
Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Müller WE, Wetter T, Suhai S (2004) Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res 14:1147–1159
Cho YG, Ishii T, Temnykh S, Chen X, Lipovich L, McCouch SR, Park WD, Ayres N, Cartinhour S (2000) Diversity of microsatellites derived from genomic libraries and GenBank sequences in rice (Oryza sativa L.). Theor Appl Genet 100:713–722
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
Crespel L, Chirollet M, Durel CE, Zhang D, Meynet J, Gudin S (2002) Mapping of qualitative and quantitative phenotypic traits in Rosa using AFLP markers. Theor Appl Genet 105:1207–1214
De Benedetti L, Burchi G, Bruna S, Mercuri A, Schiva T (2003) Use of molecular markers to improve cut flowers longevity in carnation. Acta Hort 624:343–348
Debener T, Mattiesch L (1999) Construction of a genetic linkage map for roses using RAPD and AFLP markers. Theor Appl Genet 99:891–899
Doerge RW, Churchill GA (1996) Permutation tests for multiple loci affecting a quantitative character. Genetics 142:285–294
Dugo ML, Satovic Z, Millan T, Cubero JI, Rubiales D, Cabrera A, Torres AM (2005) Genetic mapping of QTLs controlling horticultural traits in diploid roses. Theor Appl Genet 111:511–520
Dunemann F, Kahnau R, Stange I (1999) Analysis of complex leaf and flower characters in Rhododendron using a molecular linkage map. Theor Appl Genet 98:1146–1155
Ewing B, Green P (1998) Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res 8:186–194
Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res 8:175–185
Fukuoka H, Nunome T, Minamiyama Y, Kono I, Namiki N, Kojima A (2005) Read2Marker: a data processing tool for microsatellite marker development from a large data set. Biotechniques 39:472–476
Galbally J, Galbally E (1997) Carnation and pinks for garden and greenhouse. Timber Press, Portland
Hamilton RFL, Walters SM (1989) Dianthus Linnaeus. In: Walters SM, Alexander JCM, Brady A, Brickell CD, Cullen J, Green PS, Heywood VH, Matthews VA, Robson NKB, Yeo PF, Knees SG (eds) The European garden flora, vol 3. Cambridge University Press, Cambridge, pp 185–191
Hibrand-Saint Oyant L, Crespel L, Rajapakse S, Zhang L, Foucher F (2008) Genetic linkage maps of rose constructed with new microsatellite markers and locating QTL controlling flowering traits. Tree Genet Genomes 4:11–23
Kalia RK, Rai MK, Kalia S, Singh R, Dhawan AK (2011) Microsatellite markers: an overview of recent progress in plants. Euphytica 177:309–334
Kawamura K, Hibrand-Saint Oyant L, Crespel L, Thouroude T, Lalanne D, Foucher F (2011) Quantitative trait loci for flowering time and inflorescence architecture in rose. Theor Appl Genet 122:661–675
Kimura T, Yagi M, Nishitani C, Onozaki T, Ban Y, Yamamoto T (2009) Development of SSR markers in carnation (Dianthus caryophyllus). J Jpn Soc Hort Sci 78:115–123
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Lashermes P, Combes MC, Prakash NS, Trouslot P, Lorieux M, Charrier A (2001) Genetic linkage map of Coffea canephora: effect of segregation distortion and analysis of recombination rate in male and female meioses. Genome 44:589–596
Moriya S, Iwanami H, Kotoda N, Takahashi S, Yamamoto T, Abe K (2009) Development of a marker-assisted selection system for columnar growth habit in apple breeding. J Jpn Soc Hort Sci 78:279–287
Moriya S, Iwanami H, Takahashi S, Kotoda N, Suzaki K, Yamamoto T, Abe K (2010) Genetic mapping of the crown gall resistance gene of the wild apple Malus sieboldii. Tree Genet Genomes 6:195–203
Nunome T, Negoro S, Miyatake K, Yamaguchi H, Fukuoka H (2006) A protocol for the construction of microsatellite enriched genomic library. Plant Mol Biol Rep 24:305–312
Onozaki T, Yamaguchi T, Himeno M, Ikeda H (1999a) Evaluation of 277 carnation cultivars for resistance to bacterial wilt (Pseudomonas caryophylli). J Jpn Soc Hort Sci 68:546–550
Onozaki T, Yamaguchi T, Himeno M, Ikeda H (1999b) Evaluation of wild Dianthus accessions for resistance to bacterial wilt (Pseudomonas caryophylli). J Jpn Soc Hort Sci 68:974–978
Onozaki T, Ikeda H, Yamaguchi T (2001) Genetic improvement of vase life of carnation flowers by crossing and selection. Sci Hort 87:107–120
Onozaki T, Tanikawa N, Taneya M, Kudo K, Funayama T, Ikeda H, Shibata M (2004) A RAPD-derived STS marker is linked to a bacterial wilt (Burkholderia caryophylli) resistance gene in carnation. Euphytica 138:255–262
Onozaki T, Ikeda H, Shibata M, Tanikawa N, Yagi M, Yamaguchi T, Amano M (2006a) Breeding process and characteristics of Carnation Norin No. 1 ‘Miracle Rouge’ and No. 2 ‘Miracle Symphony’ with long vase life. Bull Natl Ins Flor Sci 5:1–16 (Japanese with English summary)
Onozaki T, Tanikawa N, Yagi M, Ikeda H, Sumitomo K, Shibata M (2006b) Breeding of carnations (Dianthus caryophyllus L.) for long vase life and rapid decrease in ethylene sensitivity of flowers after anthesis. J Jpn Soc Hort Sci 75:256–263
Onozaki T, Yoshinari T, Yoshimura T, Yagi M, Yoshioka S, Taneya M, Shibata M (2006c) DNA markers linked to a recessive gene controlling single flower type derived from wild species, Dianthus capitatus ssp. andrzejowskianus. Hort Res (Jpn) 5:363–367
Rajapakse S, Byrne DH, Zhang L, Anderson N, Arumuganathan K, Ballard RE (2001) Two genetic linkage maps of tetraploid roses. Theor Appl Genet 103:575–583
Rice P, Longden I, Bleasby A (2000) EMBOSS: the European molecular biology open software suite. Trends Genet 16:276–277
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, pp 365–386
Sato S, Isobe S, Asamizu E, Ohmido N, Kataoka R, Nakamura Y, Kaneko T, Sakurai N, Okumura K, Klimenko I, Sasamoto S, Wada T, Watanabe A, Kohara M, Fujishiro T, Tabata S (2005) Comprehensive structural analysis of the genome of red clover (Trifolium pratense L.). DNA Res 12:301–364
Schuelke M (2000) An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol 18:233–234
Scovel G, Ben-Meir H, Ovadis M, Itzhaki H, Vainstein A (1998) RAPD and RFLP markers tightly linked to the locus controlling carnation (Dianthus caryophyllus) flower type. Theor Appl Genet 96:117–122
Scovel G, Ovadis M, Vainstein A, Reuven M, Ben-Yephet Y (2001) Marker assisted selection for resistance to Fusarium oxysporum in the greenhouse carnation. Acta Hort 552:151–156
Smulders MJM, Rus-Kortekaas W, Vosman B (2000) Microsatellite markers useful throughout the genus Dianthus. Genome 43:208–210
Smulders MJM, Noordijk Y, Rus-Kortekaas W, Bredemeijer GMM, Vosman B (2003) Microsatellite genotyping of carnation varieties. Theor Appl Genet 106:1191–1195
Spiller M, Berger RG, Debener T (2010) Genetic dissection of scent metabolic profiles in diploid rose populations. Theor Appl Genet 120:1461–1471
Spiller M, Linde M, Hibrand-Saint Oyant L, Tsai C, Byrne D, Smulders M, Foucher F, Debener T (2011) Towards a unified genetic map for diploid roses. Theor Appl Genet 122:489–500
Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res 11:1441–1452
Vainstein A, Hillel J, Lavi U, Tzuri G (1991) Assessment of genetic relatedness in carnation by DNA fingerprint analysis. Euphytica 56:225–229
Varshney RK, Graner A, Sorrells ME (2005) Genomics-assisted breeding for crop improvement. Trends Plant Sci 10:621–630
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wang S, Basten CJ, Zeng ZB (2007). Windows QTL cartographer 2.5. Department of statistics, North Carolina State University, Raleigh. http://statgen.ncsu.edu/qtlcart/WQTLCart.htm
Wenzl P, Li H, Carling J, Zhou M, Raman H, Paul E, Hearnden P, Maier C, Xia L, Caig V, Ovesna J, Cakir M, Poulsen D, Wang J, Raman R, Smith KP, Muehlbauer GJ, Chalmers KJ, Kleinhofs A, Huttner E, Kilian A (2006) A high-density consensus map of barley linking DArT markers to SSR, RFLP and STS loci and phenotypic traits. BMC Genomics 7:206
Yagi M, Onozaki T, Taneya M, Watanabe H, Yoshimura T, Yoshinari T, Ochiai Y, Shibata M (2006a) Construction of a genetic linkage map for the carnation by using RAPD and SSR markers and mapping quantitative trait loci (QTL) for resistance to bacterial wilt caused by Burkholderia caryophylli. J Jpn Soc Hort Sci 75:166–172
Yagi M, Onozaki T, Tanikawa N, Shibata M (2006b) Molecular marker assisted selection in breeding for resistance to bacterial wilt in carnation(Dianthus caryophyllus L.). Hort Res (Jpn) 5:241–245 (Japanese with English summary)
Yagi M, Kimura T, Yamamoto T, Onozaki T (2009) Estimation of ploidy levels and breeding backgrounds in pot carnation cultivars using flow cytometry and SSR markers. J Jpn Soc Hort Sci 78:335–343
Yagi M, Onozaki T, Ikeda H, Tanikawa N, Shibata M, Yamaguchi T, Tanase K, Sumitomo K, Amano M (2010) Breeding process and characteristics of carnation ‘Karen Rouge’ with resistance to bacterial wilt. Bull Natl Inst Flor Sci 10:1–10 (Japanese with English summary)
Yan Z, Denneboom C, Hattendorf A, Dolstra O, Debener T, Stam P, Visser PB (2005) Construction of an integrated map of rose with AFLP, SSR, PK, RGA, RFLP, SCAR and morphological markers. Theor Appl Genet 110:766–777
Zeng ZB (1993) Theoretical basis of separation of multiple linked gene effects on mapping quantitative trait loci. Proc Natl Acad Sci USA 90:10972–10976
Zeng ZB (1994) Precision mapping of quantitative trait loci. Genetics 136:1457–1468
Zhang LH, Byrne DH, Ballard RE, Rajapakse S (2006) Microsatellite marker development in rose and its application in tetraploid mapping. J Am Soc Hort Sci 131:380–387
Zhang F, Chen S, Chen F, Fang W, Chen Y, Li F (2010) SRAP-based mapping and QTL detection for inflorescence-related traits in chrysanthemum (Dendranthema morifolium). Mol Breed 27:11–23. doi:10.1007/s11032-010-9409-1
Ziegle JS, Su Y, Corcoran KP, Nie L, Mayrand PE, Hoff LB, McBride LJ, Kronick MN, Diehl SR (1992) Application of automated DNA sizing technology for genotyping microsatellite loci. Genomics 14:1026–1031
Acknowledgments
This work was supported by the National Agriculture and Food Research Organization Research Project No. 211, “Establishment of Integrated Basis for Development and Application of Advanced Tools for DNA Marker-Assisted Selection in Horticultural Crops”.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yagi, M., Kimura, T., Yamamoto, T. et al. QTL analysis for resistance to bacterial wilt (Burkholderia caryophylli) in carnation (Dianthus caryophyllus) using an SSR-based genetic linkage map. Mol Breeding 30, 495–509 (2012). https://doi.org/10.1007/s11032-011-9639-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-011-9639-x