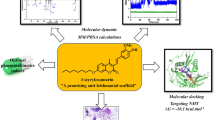
Abstract
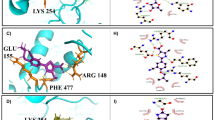
In order to combat various infectious diseases, the utilization of host-directed therapies as an alternative to chemotherapy has gained a lot of attention in the recent past, since it bypasses the existing limitations of conventional therapies. The use of host epigenetic enzymes like histone lysine methyltransferases and lysine demethylases as potential drug targets has successfully been employed for controlling various inflammatory diseases like rheumatoid arthritis and acute leukemia. In our earlier study, we have already shown that the functional knockdown of KDM6B and ASH1L in the experimental model of visceral leishmaniasis has resulted in a significant reduction of organ parasite burden. Herein, we performed a high throughput virtual screening against KDM6B and ASH1L using > 53,000 compounds that were obtained from the Maybridge library and PubChem Database, followed by molecular docking to evaluate their docking score/Glide Gscore. Based on their docking scores, the selected inhibitors were later assessed for their in vitro anti-leishmanial efficacy. Out of all inhibitors designed against KDM6B and ASH1L, HTS09796, GSK-J4 and AS-99 particularly showed promising in vitro activity with IC50 < 5 µM against both extracellular promastigote and intracellular amastigote forms of L. donovani. In vitro drug interaction studies of these inhibitors further demonstrated their synergistic interaction with amphotericin-B and miltefosine. However, GSK-J4 makes an exception by displaying an in different mode of interaction with miltefosine. Collectively, our in silico and in vitro studies acted as a platform to identify the applicability of these inhibitors targeted against KDM6B and ASH1L for anti-leishmanial therapy.
Graphical abstract










Similar content being viewed by others
Data availability
No datasets were generated or analysed during the current study.
Abbreviations
- ASH1L:
-
Histone lysine N-methyltransferase
- BLAST:
-
Basic local alignment search tool
- CC50 :
-
50% Cytotoxicity concentration
- CRK12:
-
Cell division cycle-2-related kinase 12
- DNMT:
-
DNA methyltransferases
- FDA:
-
Food and drug administration
- GUI:
-
Graphical user interface
- HDAC:
-
Histone deacetylases
- IC50 :
-
Half-maximal inhibitory concentration
- KMTs:
-
Histone lysine methyltransferases
- KDMs:
-
Histone lysine demethylases
- KDM6B:
-
Histone lysine demethylase 6B
- MTT:
-
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide
- OPLS:
-
Optimized potentials for liquid simulations
- PDB:
-
Protein data bank
- QSAR:
-
Quantitative structure- activity relationship
- RMSD:
-
Root mean square deviation
- RLU:
-
Relative luminescence unit
- VL:
-
Visceral leishmaniasis
References
Burza S, Croft SL, Boelaert M (2018) Leishmaniasis. Lancet 392:951–970. https://doi.org/10.1016/S0140-6736(18)31204-2
Uliana SRB, Trinconi CT, Coelho AC (2018) Chemotherapy of leishmaniasis: present challenges. Parasitology 145:464–480. https://doi.org/10.1017/S0031182016002523
Novais FO, Amorim CF, Scott P (2021) Host-directed therapies for cutaneous leishmaniasis. Front Immunol 12:660183. https://doi.org/10.3389/fimmu.2021.660183
van Griensven J, Balasegaram M, Meheus F et al (2010) Combination therapy for visceral leishmaniasis. Lancet Infect Dis 10:184–194. https://doi.org/10.1016/S1473-3099(10)70011-6
Martinez-Hernandez JE, Hammoud Z, de Sousa AM et al (2021) Network-based approaches reveal potential therapeutic targets for host-directed antileishmanial therapy driving drug repurposing. Microbiol Spectr 9:e0101821. https://doi.org/10.1128/Spectrum.01018-21
Takeuch O, Akira S (2011) Epigenetic control of macrophage polarization. Eur J Immunol 41:2490–2493. https://doi.org/10.1002/eji.201141792
Afrin F, Khan I, Hemeg HA (2019) Leishmania-host interactions-an epigenetic paradigm. Front Immunol 10:492. https://doi.org/10.3389/fimmu.2019.00492
Greer EL, Shi Y (2012) Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet 13:343–357. https://doi.org/10.1038/nrg3173
Tao L, Zhou Y, Luo Y et al (2024) Epigenetic regulation in cancer therapy: From mechanisms to clinical advances. MedComm Oncol 3:e59. https://doi.org/10.1002/mog2.59
Rogawski DS, Deng J, Li H et al (2021) Discovery of first-in-class inhibitors of ASH1L histone methyltransferase with anti-leukemic activity. Nat Commun 12:2792. https://doi.org/10.1038/s41467-021-23152-6
Zhao Z, Zhang Y, Gao D et al (2022) Inhibition of histone H3 lysine-27 demethylase activity relieves rheumatoid arthritis symptoms via repression of il6 transcription in macrophages. Front Immunol 13:818070. https://doi.org/10.3389/fimmu.2022.818070
Punnia-Moorthy G, Hersey P, Emran AA, Tiffen J (2021) Lysine demethylases: promising drug targets in melanoma and other cancers. Front Genet 12:680633. https://doi.org/10.3389/fgene.2021.680633
Eckschlager T, Plch J, Stiborova M, Hrabeta J (2017) Histone deacetylase inhibitors as anticancer drugs. Int J Mol Sci 18:1414. https://doi.org/10.3390/ijms18071414
Majchrzak-Celińska A, Warych A, Szoszkiewicz M (2021) Novel approaches to epigenetic therapies: from drug combinations to epigenetic editing. Genes (Basel) 12:208. https://doi.org/10.3390/genes12020208
Prieto Cárdenas LS, Arias Soler KA, Nossa González DL et al (2022) In silico antiprotozoal evaluation of 1,4-naphthoquinone derivatives against chagas and leishmaniasis diseases using QSAR, molecular docking, and ADME approaches. Pharmaceuticals (Basel) 15:687. https://doi.org/10.3390/ph15060687
Hu C, Liu X, Zeng Y et al (2021) DNA methyltransferase inhibitors combination therapy for the treatment of solid tumor: mechanism and clinical application. Clin Epigenetics 13:166. https://doi.org/10.1186/s13148-021-01154-x
Parmar N, Chandrakar P, Kar S (2020) Leishmania donovani subverts host immune response by epigenetic reprogramming of macrophage M(Lipopolysaccharides + IFN-γ)/M(IL-10) polarization. J Immunol 204:2762–2778. https://doi.org/10.4049/jimmunol.1900251
Apweiler R, Bairoch A, Wu CH et al (2004) UniProt: the universal protein knowledgebase. Nucleic Acids Res 32:D115–D119. https://doi.org/10.1093/nar/gkh131
McGinnis S, Madden TL (2004) BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Research 32: W20–W25. https://doi.org/10.1093/nar/gkh435
Boratyn GM, Camacho C, Cooper PS, et al (2013) BLAST: a more efficient report with usability improvements. Nucleic Acids Research 41: W29–W33. https://doi.org/10.1093/nar/gkt282
Goodsell DS, Zardecki C, Di Costanzo L et al (2020) RCSB protein data bank: enabling biomedical research and drug discovery. Protein Sci 29:52–65. https://doi.org/10.1002/pro.3730
Jacobson M, Sali A (2004) Comparative protein structure modeling and its applications to drug discovery. Annual reports in medicinal chemistry. Elsevier, pp 259–276
Shen MY, Sali A (2006) Statistical potential for assessment and prediction of protein structures. Protein Sci 15:2507–2524. https://doi.org/10.1110/ps.062416606
Sheik SS, Sundararajan P, Hussain ASZ, Sekar K (2002) Ramachandran plot on the web. Bioinformatics 18:1548–1549. https://doi.org/10.1093/bioinformatics/18.11.1548
Hollingsworth SA, Karplus PA (2010) A fresh look at the Ramachandran plot and the occurrence of standard structures in proteins. BioMolecular Concepts 1:271–283. https://doi.org/10.1515/bmc.2010.022
Madhavi Sastry G, Adzhigirey M, Day T et al (2013) Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des 27:221–234. https://doi.org/10.1007/s10822-013-9644-8
Kruidenier L, Chung C, Cheng Z et al (2012) A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature 488:404–408. https://doi.org/10.1038/nature11262
Michaux C, De Leval X, Julémont F et al (2006) Structure-based pharmacophore of COX-2 selective inhibitors and identification of original lead compounds from 3D database searching method. Eur J Med Chem 41:1446–1455. https://doi.org/10.1016/j.ejmech.2006.07.017
Devi B, Rajagopal K, Elizabeth E (2017) Pharmacophoric screening of various endophytic fungal metabolites. Asian J Pharm Clin Res 10:140. https://doi.org/10.22159/ajpcr.2017.v10i5.17157
Ribeiro R, Botelho FD, Pinto AMV et al (2023) Molecular modeling study of natural products as potential bioactive compounds against SARS-CoV-2. J Mol Model 29:183. https://doi.org/10.1007/s00894-023-05586-5
Dixon SL, Smondyrev AM, Knoll EH et al (2006) PHASE: a new engine for pharmacophore perception, 3D QSAR model development, and 3D database screening: 1. Methodology and preliminary results. J Comput Aided Mol Des 20:647–671. https://doi.org/10.1007/s10822-006-9087-6
Pribadi C, Camp E, Cakouros D et al (2020) Pharmacological targeting of KDM6A and KDM6B, as a novel therapeutic strategy for treating craniosynostosis in Saethre-Chotzen syndrome. Stem Cell Res Ther 11:529. https://doi.org/10.1186/s13287-020-02051-5
Hung PH, Hsu YC, Chen TH et al (2022) The histone demethylase inhibitor GSK-J4 Is a therapeutic target for the kidney fibrosis of diabetic kidney disease via DKK1 modulation. Int J Mol Sci 23:9407. https://doi.org/10.3390/ijms23169407
Bai SB, Geethavani M, Ramakrishna C (2022) Synthesis characterization and molinspiration analysis, anti-bacterial activity of novel 2,4,6-tri substituted pyrimidines. J Young Pharm 14:174–178. https://doi.org/10.5530/jyp.2022.14.33
Ntie-Kang F (2013) An in silico evaluation of the ADMET profile of the StreptomeDB database. Springerplus 2:353. https://doi.org/10.1186/2193-1801-2-353
Hou P, Huang C, Liu C-P et al (2019) Structural insights into stimulation of Ash1L’s H3K36 methyltransferase activity through Mrg15 binding. Structure 27:837–845.e3. https://doi.org/10.1016/j.str.2019.01.015
Friesner RA, Murphy RB, Repasky MP et al (2006) Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein−ligand complexes. J Med Chem 49:6177–6196. https://doi.org/10.1021/jm051256o
Anand D, Yadav PK, Patel OPS et al (2017) Antileishmanial activity of pyrazolopyridine derivatives and their potential as an adjunct therapy with miltefosine. J Med Chem 60:1041–1059. https://doi.org/10.1021/acs.jmedchem.6b01447
Huber W, Koella JC (1993) A comparison of three methods of estimating EC50 in studies of drug resistance of malaria parasites. Acta Trop 55:257–261. https://doi.org/10.1016/0001-706x(93)90083-n
Fivelman QL, Adagu IS, Warhurst DC (2004) Modified fixed-ratio isobologram method for studying in vitro interactions between atovaquone and proguanil or dihydroartemisinin against drug-resistant strains of Plasmodium falciparum. Antimicrob Agents Chemother 48:4097–4102. https://doi.org/10.1128/AAC.48.11.4097-4102.2004
De I, Müller CW (2019) Unleashing the power of ASH1L methyltransferase. Structure 27:727–728. https://doi.org/10.1016/j.str.2019.04.012
Yu M, Jia Y, Ma Z et al (2022) Structural insight into ASH1L PHD finger recognizing methylated histone H3K4 and promoting cell growth in prostate cancer. Front Oncol 12:906807. https://doi.org/10.3389/fonc.2022.906807
Kurmasheva RT, Erickson SW, Han R et al (2021) In vivo evaluation of the lysine-specific demethylase (KDM1A/LSD1) inhibitor SP-2577 (Seclidemstat) against pediatric sarcoma preclinical models: a report from the pediatric preclinical testing consortium (PPTC). Pediatr Blood Cancer 68:e29304. https://doi.org/10.1002/pbc.29304
Reed DR, Mascarenhas L, Meyers PA et al (2020) A phase I/II clinical trial of the reversible LSD1 inhibitor, seclidemstat, in patients with relapsed/refractory Ewing sarcoma. JCO 38:TPS11567–TPS11567. https://doi.org/10.1200/JCO.2020.38.15_suppl.TPS11567
Ahmad S, Bhagwati S, Kumar S et al (2020) Molecular modeling assisted identification and biological evaluation of potent cathepsin S inhibitors. J Mol Graph Model 96:107512. https://doi.org/10.1016/j.jmgm.2019.107512
Jorgensen WL, Maxwell DS, Tirado-Rives J (1996) Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc 118:11225–11236. https://doi.org/10.1021/ja9621760
Bhagwati S, Siddiqi MI (2020) Identification of potential soluble epoxide hydrolase (sEH) inhibitors by ligand-based pharmacophore model and biological evaluation. J Biomol Struct Dyn 38:4956–4966. https://doi.org/10.1080/07391102.2019.1691659
Rebello KM, Andrade-Neto VV, Gomes CRB et al (2019) Miltefosine-lopinavir combination therapy against Leishmania infantum infection: in vitro and in vivo approaches. Front Cell Infect Microbiol 9:229. https://doi.org/10.3389/fcimb.2019.00229
Emiliano YSS, Almeida-Amaral EE (2018) Efficacy of apigenin and miltefosine combination therapy against experimental Cutaneous Leishmaniasis. J Nat Prod 81:1910–1913. https://doi.org/10.1021/acs.jnatprod.8b00356
Berenbaum MC (1989) What is synergy? Pharmacol Rev 41:93–141
Lobo-Silva J, Cabral FJ, Amaral MS et al (2020) The antischistosomal potential of GSK-J4, an H3K27 demethylase inhibitor: insights from molecular modeling, transcriptomics and in vitro assays. Parasit Vectors 13:140. https://doi.org/10.1186/s13071-020-4000-z
Kashif M, Paladhi A, Singh R et al (2020) Leishmanicidal activity of an in silico-screened novel inhibitor against ascorbate peroxidase of Leishmania donovani. Antimicrob Agents Chemother 64:e01766–e01819. https://doi.org/10.1128/AAC.01766-19
Ângelo de Souza L, Silva Bastos M, de Melo AJ et al (2020) Histone deacetylases inhibitors as new potential drugs against Leishmania braziliensis, the main causative agent of new world tegumentary leishmaniasis. Biochem Pharmacol 180:114191. https://doi.org/10.1016/j.bcp.2020.114191
Huang L, Jiang Y, Chen Y (2017) Predicting drug combination index and simulating the network-regulation dynamics by mathematical modeling of drug-targeted EGFR-ERK signaling pathway. Sci Rep 7:40752. https://doi.org/10.1038/srep40752
Huang RY, Pei L, Liu Q et al (2019) Isobologram analysis: a comprehensive review of methodology and current research. Front Pharmacol 10:1222. https://doi.org/10.3389/fphar.2019.01222
Acknowledgements
The authors would like to thank Dr. Neena Goyal, Division of Biochemistry and Structural Biology, CSIR-CDRI, Lucknow, India, for providing us with transgenic L. donovani promastigotes. The authors extend their gratitude to Dr. Sanjay Batra, Division of Medicinal and Process Chemistry, CSIR-CDRI, Lucknow, India, for providing the Maybridge library compounds (sourced from the chemical repository of CSIR-CDRI, Lucknow) for the in vitro assessment conducted in this study.
Funding
The study was funded by the Department of Biotechnology (BT/PR32490/MED/29/1457/2019) & Department of Science & Technology (DST, CRG/2020/002932), New Delhi, India and CSIR-CDRI in-house project. The author M.D. & U.K. is thankful to CSIR & UGC, New Delhi respectively for providing financial assistance in the form of fellowship. T.Q. acknowledges the Department of Science & Technology (DST, CRG/2020/002932), New Delhi, for the project fellowship.
Author information
Authors and Affiliations
Contributions
MD, TQ and SK conceived and designed the study. MD and TQ performed and analyzed the results of biological experiments. TQ, UK and MIS performed and analyzed the results of computational studies. MD and TQ prepared the figures and wrote the original manuscript. Overall supervision, conceptualization, acquisition of funding & manuscript editing was done by SK. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors state that none of the work described in this manuscript appears to have been influenced by any known financial interests or personal relationships.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dutta, M., Qamar, T., Kushavah, U. et al. Exploring host epigenetic enzymes as targeted therapies for visceral leishmaniasis: in silico design and in vitro efficacy of KDM6B and ASH1L inhibitors. Mol Divers (2024). https://doi.org/10.1007/s11030-024-10824-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11030-024-10824-w